Disruption of the histone acetyltransferase MYST4 leads to a Noonan syndrome-like phenotype and hyperactivated MAPK signaling in humans and mice
- PMID: 21804188
- PMCID: PMC3163944
- DOI: 10.1172/JCI43428
Disruption of the histone acetyltransferase MYST4 leads to a Noonan syndrome-like phenotype and hyperactivated MAPK signaling in humans and mice
Abstract
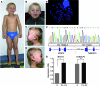
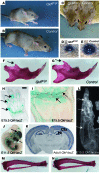
Epigenetic regulation of gene expression, through covalent modification of histones, is a key process controlling growth and development. Accordingly, the transcription factors regulating these processes are important targets of genetic diseases. However, surprisingly little is known about the relationship between aberrant epigenetic states, the cellular process affected, and their phenotypic consequences. By chromosomal breakpoint mapping in a patient with a Noonan syndrome-like phenotype that encompassed short stature, blepharoptosis, and attention deficit hyperactivity disorder, we identified haploinsufficiency of the histone acetyltransferase gene MYST histone acetyltransferase (monocytic leukemia) 4 (MYST4), as the underlying cause of the phenotype. Using acetylation, whole genome expression, and ChIP studies in cells from the patient, cell lines in which MYST4 expression was knocked down using siRNA, and the Myst4 querkopf mouse, we found that H3 acetylation is important for neural, craniofacial, and skeletal morphogenesis, mainly through its ability to specifically regulating the MAPK signaling pathway. This finding further elucidates the complex role of histone modifications in mammalian development and adds what we believe to be a new mechanism to the pathogenic phenotypes resulting from misregulation of the RAS signaling pathway.
Figures





References
-
- MacDonald VE, Howe LJ. Histone acetylation: where to go and how to get there. Epigenetics. 2009;4(3):139–143. - PubMed

