Analysis of imprinted gene expression in normal fertilized and uniparental preimplantation porcine embryos
- PMID: 21804912
- PMCID: PMC3137617
- DOI: 10.1371/journal.pone.0022216
Analysis of imprinted gene expression in normal fertilized and uniparental preimplantation porcine embryos
Abstract
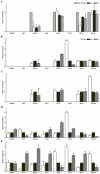
In the present study quantitative real-time PCR was used to determine the expression status of eight imprinted genes (GRB10, H19, IGF2R, XIST, IGF2, NNAT, PEG1 and PEG10) during preimplantation development, in normal fertilized and uniparental porcine embryos. The results demonstrated that, in all observed embryo samples, a non imprinted gene expression pattern up to the 16-cell stage of development was common for most genes. This was true for all classes of embryo, regardless of parental-origins and the direction of imprint. However, several differentially expressed genes (H19, IGF2, XIST and PEG10) were detected amongst the classes at the blastocyst stage of development. Most interestingly and despite the fact that maternally and paternally expressed genes should not be expressed in androgenones and parthenogenones, respectively, both uniparental embryos expressed these genes when tested for in this study. In order to account for this phenomenon, we compared the expression patterns of eight imprinted genes along with the methylation status of the IGF2/H19 DMR3 in haploid and diploid parthenogenetic embryos. Our findings revealed that IGF2, NNAT and PEG10 were silenced in haploid but not diploid parthenogenetic blastocysts and differential methylation of the IGF2/H19 DMR3 was consistently observed between haploid and diploid parthenogenetic blastocysts. These results appear to suggest that there exists a process to adjust the expression status of imprinted genes in diploid parthenogenetic embryos and that this phenomenon may be associated with altered methylation at an imprinting control region. In addition we believe that imprinted expression occurs in at least four genes, namely H19, IGF2, XIST and PEG10 in porcine blastocyst stage embryos.
Conflict of interest statement
Figures
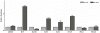
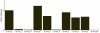
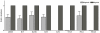

Similar articles
-
Amino acid supplementation affects imprinted gene transcription patterns in parthenogenetic porcine blastocysts.PLoS One. 2014 Sep 2;9(9):e106549. doi: 10.1371/journal.pone.0106549. eCollection 2014. PLoS One. 2014. PMID: 25180972 Free PMC article.
-
Monoallelic expression of nine imprinted genes in the sheep embryo occurs after the blastocyst stage.Reproduction. 2008 Jan;135(1):29-40. doi: 10.1530/REP-07-0211. Reproduction. 2008. PMID: 18159081
-
Analysis of imprinted IGF2/H19 gene methylation and expression in normal fertilized and parthenogenetic embryonic stem cells of pigs.Anim Reprod Sci. 2014 Jun 10;147(1-2):47-55. doi: 10.1016/j.anireprosci.2014.03.020. Epub 2014 Apr 12. Anim Reprod Sci. 2014. PMID: 24794444
-
Imprinting of the mouse Igf2r gene depends on an intronic CpG island.Mol Cell Endocrinol. 1998 May 25;140(1-2):9-14. doi: 10.1016/s0303-7207(98)00022-7. Mol Cell Endocrinol. 1998. PMID: 9722161 Review.
-
Expression of imprinted genes in human preimplantation development.Mol Cell Endocrinol. 2001 Oct 22;183 Suppl 1:S35-40. doi: 10.1016/s0303-7207(01)00575-5. Mol Cell Endocrinol. 2001. PMID: 11576730 Review.
Cited by
-
H19/Igf2 Expression and Methylation of Histone 3 in Mice Chimeric Blastocysts.Rep Biochem Mol Biol. 2020 Oct;9(3):357-365. doi: 10.29252/rbmb.9.3.357. Rep Biochem Mol Biol. 2020. PMID: 33649730 Free PMC article.
-
The perturbed expression of m6A in parthenogenetic mouse embryos.Genet Mol Biol. 2019 Jul-Sep;42(3):666-670. doi: 10.1590/1678-4685-GMB-2018-0212. Epub 2019 Nov 14. Genet Mol Biol. 2019. PMID: 31188932 Free PMC article.
-
Karyotype characterization of in vivo- and in vitro-derived porcine parthenogenetic cell lines.PLoS One. 2014 May 20;9(5):e97974. doi: 10.1371/journal.pone.0097974. eCollection 2014. PLoS One. 2014. PMID: 24844788 Free PMC article.
-
Genome-wide identification of imprinted genes in pigs and their different imprinting status compared with other mammals.Zool Res. 2020 Nov 18;41(6):721-725. doi: 10.24272/j.issn.2095-8137.2020.072. Zool Res. 2020. PMID: 32808516 Free PMC article.
-
Genome wide screening of candidate genes for improving piglet birth weight using high and low estimated breeding value populations.Int J Biol Sci. 2014 Feb 7;10(3):236-44. doi: 10.7150/ijbs.7744. eCollection 2014. Int J Biol Sci. 2014. PMID: 24644423 Free PMC article.
References
-
- Kelsey G, Reik W. Analysis and identification of imprinted genes. Methods. 1998;14:211–234. - PubMed
-
- Barton SC, Surani MA, Norris ML. Role of paternal and maternal genomes in mouse development. Nature. 1984;311:374–376. - PubMed
-
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. - PubMed
-
- Khosla S, Dean W, Reik W, Feil R. Culture of preimplantation embryos and its long-term effects on gene expression and phenotype. Hum Reprod Update. 2001;7:419–427. - PubMed
-
- Mann MR, Lee SS, Doherty AS, Verona RI, Nolen LD, et al. Selective loss of imprinting in the placenta following preimplantation development in culture. Development. 2004;131:3727–3735. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

