The 2nd DBCLS BioHackathon: interoperable bioinformatics Web services for integrated applications
- PMID: 21806842
- PMCID: PMC3170566
- DOI: 10.1186/2041-1480-2-4
The 2nd DBCLS BioHackathon: interoperable bioinformatics Web services for integrated applications
Abstract
Background: The interaction between biological researchers and the bioinformatics tools they use is still hampered by incomplete interoperability between such tools. To ensure interoperability initiatives are effectively deployed, end-user applications need to be aware of, and support, best practices and standards. Here, we report on an initiative in which software developers and genome biologists came together to explore and raise awareness of these issues: BioHackathon 2009.
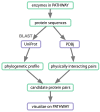
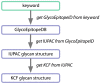
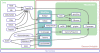
Results: Developers in attendance came from diverse backgrounds, with experts in Web services, workflow tools, text mining and visualization. Genome biologists provided expertise and exemplar data from the domains of sequence and pathway analysis and glyco-informatics. One goal of the meeting was to evaluate the ability to address real world use cases in these domains using the tools that the developers represented. This resulted in i) a workflow to annotate 100,000 sequences from an invertebrate species; ii) an integrated system for analysis of the transcription factor binding sites (TFBSs) enriched based on differential gene expression data obtained from a microarray experiment; iii) a workflow to enumerate putative physical protein interactions among enzymes in a metabolic pathway using protein structure data; iv) a workflow to analyze glyco-gene-related diseases by searching for human homologs of glyco-genes in other species, such as fruit flies, and retrieving their phenotype-annotated SNPs.
Conclusions: Beyond deriving prototype solutions for each use-case, a second major purpose of the BioHackathon was to highlight areas of insufficiency. We discuss the issues raised by our exploration of the problem/solution space, concluding that there are still problems with the way Web services are modeled and annotated, including: i) the absence of several useful data or analysis functions in the Web service "space"; ii) the lack of documentation of methods; iii) lack of compliance with the SOAP/WSDL specification among and between various programming-language libraries; and iv) incompatibility between various bioinformatics data formats. Although it was still difficult to solve real world problems posed to the developers by the biological researchers in attendance because of these problems, we note the promise of addressing these issues within a semantic framework.
Figures



References
-
- Database Center for Life Science. http://dbcls.rois.ac.jp/
-
- Katayama T, Arakawa K, Nakao M, Ono K, Aoki-Kinoshita K, Yamamoto Y, Yamaguchi A, Kawashima S, Chun H-W, Aerts J. et al.The DBCLS BioHackathon: standardization and interoperability for bioinformatics web services and workflows. Journal of biomedical semantics. 2010;1:8. doi: 10.1186/2041-1480-1-8. - DOI - PMC - PubMed
-
- Okinawa Institute of Science and Technology. http://www.oist.jp/
-
- Web API for Biology (WABI) http://xml.nig.ac.jp/index.html

