Autophosphorylation in the leucine-rich repeat kinase 2 (LRRK2) GTPase domain modifies kinase and GTP-binding activities
- PMID: 21806997
- PMCID: PMC3158845
- DOI: 10.1016/j.jmb.2011.07.033
Autophosphorylation in the leucine-rich repeat kinase 2 (LRRK2) GTPase domain modifies kinase and GTP-binding activities
Erratum in
-
Corrigendum to "Autophosphorylation in the Leucine-Rich Repeat Kinase 2 (LRRK2) GTPase Domain Modified Kinase and GTP-binding Activities" [J. Mol. Biol. 412(1) (2011) 94-110].J Mol Biol. 2021 Sep 3;433(18):167129. doi: 10.1016/j.jmb.2021.167129. Epub 2021 Jul 20. J Mol Biol. 2021. PMID: 34294467 Free PMC article. No abstract available.
Abstract
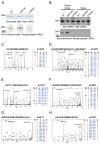
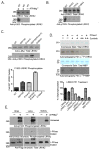
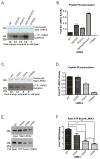
The leucine-rich repeat kinase 2 (LRRK2) protein has both guanosine triphosphatase (GTPase) and kinase activities, and mutation in either enzymatic domain can cause late-onset Parkinson disease. Nucleotide binding in the GTPase domain may be required for kinase activity, and residues in the GTPase domain are potential sites for autophosphorylation, suggesting a complex mechanism of intrinsic regulation. To further define the effects of LRRK2 autophosphorylation, we applied a technique optimal for detection of protein phosphorylation, electron transfer dissociation, and identified autophosphorylation events exclusively nearby the nucleotide binding pocket in the GTPase domain. Parkinson-disease-linked mutations alter kinase activity but did not alter autophosphorylation site specificity or sites of phosphorylation in a robust in vitro substrate myelin basic protein. Amino acid substitutions in the GTPase domain have large effects on kinase activity, as insertion of the GTPase-associated R1441C pathogenic mutation together with the G2019S kinase domain mutation resulted in a multiplicative increase (∼7-fold) in activity. Removal of a conserved autophosphorylation site (T1503) by mutation to an alanine residue resulted in greatly decreased GTP-binding and kinase activities. While autophosphorylation likely serves to potentiate kinase activity, we find that oligomerization and loss of the active dimer species occur in an ATP- and autophosphorylation-independent manner. LRRK2 autophosphorylation sites are overall robustly protected from dephosphorylation in vitro, suggesting tight control over activity in vivo. We developed highly specific antibodies targeting pT1503 but failed to detect endogenous autophosphorylation in protein derived from transgenic mice and cell lines. LRRK2 activity in vivo is unlikely to be constitutive but rather refined to specific responses.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures



Similar articles
-
The Parkinson's disease kinase LRRK2 autophosphorylates its GTPase domain at multiple sites.Biochem Biophys Res Commun. 2009 Nov 20;389(3):449-54. doi: 10.1016/j.bbrc.2009.08.163. Epub 2009 Sep 3. Biochem Biophys Res Commun. 2009. PMID: 19733152 Free PMC article.
-
The dual enzyme LRRK2 hydrolyzes GTP in both its GTPase and kinase domains in vitro.Biochim Biophys Acta Proteins Proteom. 2017 Mar;1865(3):274-280. doi: 10.1016/j.bbapap.2016.12.001. Epub 2016 Dec 8. Biochim Biophys Acta Proteins Proteom. 2017. PMID: 27939437 Free PMC article.
-
GTPase activity regulates kinase activity and cellular phenotypes of Parkinson's disease-associated LRRK2.Hum Mol Genet. 2013 Mar 15;22(6):1140-56. doi: 10.1093/hmg/dds522. Epub 2012 Dec 13. Hum Mol Genet. 2013. PMID: 23241358
-
Contribution of GTPase activity to LRRK2-associated Parkinson disease.Small GTPases. 2013 Jul-Sep;4(3):164-70. doi: 10.4161/sgtp.25130. Epub 2013 Jun 10. Small GTPases. 2013. PMID: 24025585 Free PMC article. Review.
-
Understanding the GTPase Activity of LRRK2: Regulation, Function, and Neurotoxicity.Adv Neurobiol. 2017;14:71-88. doi: 10.1007/978-3-319-49969-7_4. Adv Neurobiol. 2017. PMID: 28353279 Free PMC article. Review.
Cited by
-
The cell biology of Parkinson's disease.J Cell Biol. 2021 Apr 5;220(4):e202012095. doi: 10.1083/jcb.202012095. J Cell Biol. 2021. PMID: 33749710 Free PMC article. Review.
-
Unique functional and structural properties of the LRRK2 protein ATP-binding pocket.J Biol Chem. 2014 Nov 21;289(47):32937-51. doi: 10.1074/jbc.M114.602318. Epub 2014 Sep 16. J Biol Chem. 2014. PMID: 25228699 Free PMC article.
-
GTP binding and intramolecular regulation by the ROC domain of Death Associated Protein Kinase 1.Sci Rep. 2012;2:695. doi: 10.1038/srep00695. Epub 2012 Sep 26. Sci Rep. 2012. PMID: 23019516 Free PMC article.
-
LRRK2 phosphorylates pre-synaptic N-ethylmaleimide sensitive fusion (NSF) protein enhancing its ATPase activity and SNARE complex disassembling rate.Mol Neurodegener. 2016 Jan 13;11:1. doi: 10.1186/s13024-015-0066-z. Mol Neurodegener. 2016. PMID: 26758690 Free PMC article.
-
LRRK2 kinase activity and biology are not uniformly predicted by its autophosphorylation and cellular phosphorylation site status.Front Mol Neurosci. 2014 Jun 24;7:54. doi: 10.3389/fnmol.2014.00054. eCollection 2014. Front Mol Neurosci. 2014. PMID: 25009464 Free PMC article.
References
-
- Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, Lopez de Munain A, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Carrera IM, Pena AS, de Silva R, Lees A, Marti-Masso JF, Perez-Tur J, Wood NW, Singleton AB. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44:595–600. - PubMed
-
- Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, Stoessl AJ, Pfeiffer RF, Patenge N, Carbajal IC, Vieregge P, Asmus F, Muller-Myhsok B, Dickson DW, Meitinger T, Strom TM, Wszolek ZK, Gasser T. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–7. - PubMed
-
- West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, Biskup S, Zhang Z, Lim KL, Dawson VL, Dawson TM. Parkinson’s disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet. 2007;16:223–32. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

