The role of FACT in making and breaking nucleosomes
- PMID: 21807128
- PMCID: PMC3229669
- DOI: 10.1016/j.bbagrm.2011.07.009
The role of FACT in making and breaking nucleosomes
Abstract
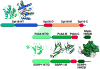
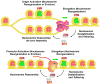
FACT is a roughly 180kDa heterodimeric protein complex important for managing the properties of chromatin in eukaryotic cells. Chromatin is a repressive barrier that plays an important role in protecting genomic DNA and regulating access to it. This barrier must be temporarily removed during transcription, replication, and repair, but it also must be rapidly restored to the original state afterwards. Further, the properties of chromatin are dynamic and must be adjusted as conditions dictate. FACT was identified as a factor that destabilizes nucleosomes in vitro, but it has now also been implicated as a central factor in the deposition of histones to form nucleosomes, as an exchange factor that swaps the histones within existing nucleosomes for variant forms, and as a tether that prevents histones from being displaced by the passage of RNA polymerases during transcription. FACT therefore plays central roles in building, maintaining, adjusting, and overcoming the chromatin barrier. This review summarizes recent results that have begun to reveal how FACT can promote what appear to be contradictory goals, using a simple set of binding activities to both enhance and diminish the stability of nucleosomes. This article is part of a Special Issue entitled: Histone chaperones and Chromatin assembly.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures
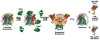
References
-
- Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. - PubMed
-
- Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises the human SPT16/CDC68 and SSRP1 proteins. Nature. 1999;400:284–288. - PubMed
-
- Reinberg D, Sims RJ., 3rd de FACTo nucleosome dynamics. J Biol Chem. 2006;281:23297–23301. - PubMed
-
- Brewster NK, Johnston GC, Singer RA. Characterization of the CP complex, an abundant dimer of Cdc68 and Pob3 proteins that regulates yeast transcriptional activation and chromatin repression. J Biol Chem. 1998;273:21972–21979. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

