PARP regulates nonhomologous end joining through retention of Ku at double-strand breaks
- PMID: 21807880
- PMCID: PMC3153639
- DOI: 10.1083/jcb.201012132
PARP regulates nonhomologous end joining through retention of Ku at double-strand breaks
Abstract
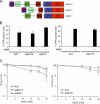
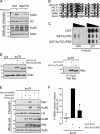
Poly adenosine diphosphate (ADP)-ribosylation (PARylation) by poly ADP-ribose (PAR) polymerases (PARPs) is an early response to DNA double-strand breaks (DSBs). In this paper, we exploit Dictyostelium discoideum to uncover a novel role for PARylation in regulating nonhomologous end joining (NHEJ). PARylation occurred at single-strand breaks, and two PARPs, Adprt1b and Adprt2, were required for resistance to this kind of DNA damage. In contrast, although Adprt1b was dispensable for PARylation at DSBs, Adprt1a and, to a lesser extent, Adprt2 were required for this event. Disruption of adprt2 had a subtle impact on the ability of cells to perform NHEJ. However, disruption of adprt1a decreased the ability of cells to perform end joining with a concomitant increase in homologous recombination. PAR-dependent regulation of NHEJ was achieved through promoting recruitment and/or retention of Ku at DSBs. Furthermore, a PAR interaction motif in Ku70 was required for this regulation and efficient NHEJ. These data illustrate that PARylation at DSBs promotes NHEJ through recruitment or retention of repair factors at sites of DNA damage.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

