Human CABIN1 is a functional member of the human HIRA/UBN1/ASF1a histone H3.3 chaperone complex
- PMID: 21807893
- PMCID: PMC3187368
- DOI: 10.1128/MCB.05546-11
Human CABIN1 is a functional member of the human HIRA/UBN1/ASF1a histone H3.3 chaperone complex
Abstract
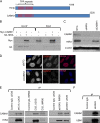
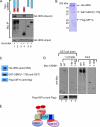
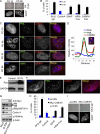
The mammalian HIRA/UBN1/ASF1a complex is a histone chaperone complex that is conserved from yeast (Saccharomyces cerevisiae) to humans. This complex preferentially deposits the histone variant H3.3 into chromatin in a DNA replication-independent manner and is implicated in diverse chromatin regulatory events from gene activation to heterochromatinization. In yeast, the orthologous complex consists of three Hir proteins (Hir1p, Hir2p, and Hir3p), Hpc2p, and Asf1p. Yeast Hir3p has weak homology to CABIN1, a fourth member of the human complex, suggesting that Hir3p and CABIN1 may be orthologs. Here we show that HIRA and CABIN1 interact at ectopic and endogenous levels of expression in cells, and we isolate the quaternary HIRA/UBN1/CABIN1/ASF1a (HUCA) complex, assembled from recombinant proteins. Mutational analyses support the view that HIRA acts as a scaffold to bring together UBN1, ASF1a, and CABIN1 into a quaternary complex. We show that, like HIRA, UBN1, and ASF1a, CABIN1 is involved in heterochromatinization of the genome of senescent human cells. Moreover, in proliferating cells, HIRA and CABIN1 regulate overlapping sets of genes, and these genes are enriched in the histone variant H3.3. In sum, these data demonstrate that CABIN1 is a functional member of the human HUCA complex and so is the likely ortholog of yeast Hir3p.
Figures



References
-
- Adams P. D. 2009. Healing and hurting: molecular mechanisms, functions, and pathologies of cellular senescence. Mol. Cell 36:2–14 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
