Dolutegravir (S/GSK1349572) exhibits significantly slower dissociation than raltegravir and elvitegravir from wild-type and integrase inhibitor-resistant HIV-1 integrase-DNA complexes
- PMID: 21807982
- PMCID: PMC3187001
- DOI: 10.1128/AAC.00157-11
Dolutegravir (S/GSK1349572) exhibits significantly slower dissociation than raltegravir and elvitegravir from wild-type and integrase inhibitor-resistant HIV-1 integrase-DNA complexes
Abstract
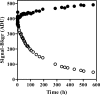
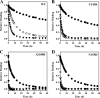
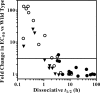
The integrase inhibitor (INI) dolutegravir (DTG; S/GSK1349572) has significant activity against HIV-1 isolates with raltegravir (RAL)- and elvitegravir (ELV)-associated resistance mutations. As an initial step in characterizing the different resistance profiles of DTG, RAL, and ELV, we determined the dissociation rates of these INIs with integrase (IN)-DNA complexes containing a broad panel of IN proteins, including IN substitutions corresponding to signature RAL and ELV resistance mutations. DTG dissociates slowly from a wild-type IN-DNA complex at 37°C with an off-rate of 2.7 × 10(-6) s(-1) and a dissociative half-life (t(1/2)) of 71 h, significantly longer than the half-lives for RAL (8.8 h) and ELV (2.7 h). Prolonged binding (t(1/2), at least 5 h) was observed for DTG with IN-DNA complexes containing E92, Y143, Q148, and N155 substitutions. The addition of a second substitution to either Q148 or N155 typically resulted in an increase in the off-rate compared to that with the single substitution. For all of the IN substitutions tested, the off-rate of DTG from IN-DNA complexes was significantly slower (from 5 to 40 times slower) than the off-rate of RAL or ELV. These data are consistent with the potential for DTG to have a higher genetic barrier to resistance, provide evidence that the INI off-rate may be an important component of the mechanism of INI resistance, and suggest that the slow dissociation of DTG may contribute to its distinctive resistance profile.
Figures
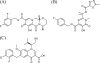

References
-
- Asante-Appiah E., Skalka A. M. 1999. HIV-1 integrase: structural organization, conformational changes, and catalysis. Adv. Virus Res. 52:351–369 - PubMed
-
- Boros E. E., Johns B. A., Garvey E. P., Koble C. S., Miller W. H. 2006. Synthesis and HIV-integrase strand transfer inhibition activity of 7-hyroxyl[1,3]thiazolo[5,4-b]pyridin-5(4H)-ones. Bioorg. Med. Chem. Lett. 16:5668–5672 - PubMed
-
- Ceccherini-Silberstein F., et al. 2009. Characterization and structural analysis of HIV-1 integrase conservation. AIDS Rev. 11:17–29 - PubMed
-
- Chiu T. K., Davies D. R. 2004. Structure and function of HIV-1 integrase. Curr. Top. Med. Chem. 4:965–977 - PubMed
-
- Cooper D. A., et al. 2008. Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N. Engl. J. Med. 359:355–365 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

