Genetic framework for GATA factor function in vascular biology
- PMID: 21808000
- PMCID: PMC3158141
- DOI: 10.1073/pnas.1108440108
Genetic framework for GATA factor function in vascular biology
Abstract
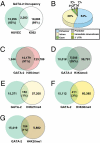
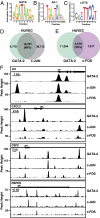
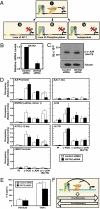
Vascular endothelial dysfunction underlies the genesis and progression of numerous diseases. Although the GATA transcription factor GATA-2 is expressed in endothelial cells and is implicated in coronary heart disease, it has been studied predominantly as a master regulator of hematopoiesis. Because many questions regarding GATA-2 function in the vascular biology realm remain unanswered, we used ChIP sequencing and loss-of-function strategies to define the GATA-2-instigated genetic network in human endothelial cells. In contrast to erythroid cells, GATA-2 occupied a unique target gene ensemble consisting of genes encoding key determinants of endothelial cell identity and inflammation. GATA-2-occupied sites characteristically contained motifs that bind activator protein-1 (AP-1), a pivotal regulator of inflammatory genes. GATA-2 frequently occupied the same chromatin sites as c-JUN and c-FOS, heterodimeric components of AP-1. Although all three components were required for maximal AP-1 target gene expression, GATA-2 was not required for AP-1 chromatin occupancy. GATA-2 conferred maximal phosphorylation of chromatin-bound c-JUN at Ser-73, which stimulates AP-1-dependent transactivation, in a chromosomal context-dependent manner. This work establishes a link between a GATA factor and inflammatory genes, mechanistic insights underlying GATA-2-AP-1 cooperativity and a rigorous genetic framework for understanding GATA-2 function in normal and pathophysiological vascular states.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Tsai F-Y, Orkin SH. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 1997;89:3636–3643. - PubMed
-
- Tsai FY, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. - PubMed
-
- Dorfman DM, Wilson DB, Bruns GA, Orkin SH. Human transcription factor GATA-2. Evidence for regulation of preproendothelin-1 gene expression in endothelial cells. J Biol Chem. 1992;267:1279–1285. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

