Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue
- PMID: 21808026
- PMCID: PMC3158156
- DOI: 10.1073/pnas.1108161108
Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue
Abstract
We describe a label-free imaging method to monitor stem-cell metabolism that discriminates different states of stem cells as they differentiate in living tissues. In this method we use intrinsic fluorescence biomarkers and the phasor approach to fluorescence lifetime imaging microscopy in conjunction with image segmentation, which we use to introduce the concept of the cell phasor. In live tissues we are able to identify intrinsic fluorophores, such as collagen, retinol, retinoic acid, porphyrin, flavins, and free and bound NADH. We have exploited the cell phasor approach to detect a trend in metabolite concentrations along the main axis of the Caenorhabditis elegans germ line. This trend is consistent with known changes in metabolic states during differentiation. The cell phasor approach to lifetime imaging provides a label-free, fit-free, and sensitive method to identify different metabolic states of cells during differentiation, to sense small changes in the redox state of cells, and may identify symmetric and asymmetric divisions and predict cell fate. Our method is a promising noninvasive optical tool for monitoring metabolic pathways during differentiation or disease progression, and for cell sorting in unlabeled tissues.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
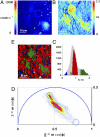
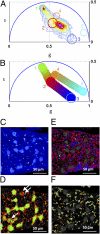
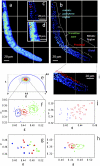
 ) corresponds to the 64 levels of the contours that indicate the percent occurrence in the phasor histogram of the pixels of the image. Four clusters corresponding to different tissue components are identified in the phasor distribution with different colors. (E) Phasor color maps of the FLIM image. The colors of pixels correspond to the clusters of tissue components selected in the phasor plot.
) corresponds to the 64 levels of the contours that indicate the percent occurrence in the phasor histogram of the pixels of the image. Four clusters corresponding to different tissue components are identified in the phasor distribution with different colors. (E) Phasor color maps of the FLIM image. The colors of pixels correspond to the clusters of tissue components selected in the phasor plot.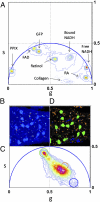
References
-
- Donovan PJ, Gearhart J. The end of the beginning for pluripotent stem cells. Nature. 2001;414(6859):92–97. - PubMed
-
- Knoblich JA. Asymmetric cell division during animal development. Nat Rev Mol Cell Biol. 2001;2(1):11–20. - PubMed
-
- Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. - PubMed
-
- Pera MF, Tam PP. Extrinsic regulation of pluripotent stem cells. Nature. 2010;465:713–720. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

