Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA
- PMID: 21808029
- PMCID: PMC3158146
- DOI: 10.1073/pnas.1109263108
Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA
Abstract
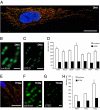
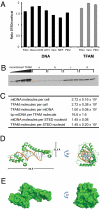
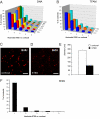
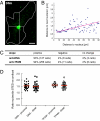
Mammalian mtDNA is packaged in DNA-protein complexes denoted mitochondrial nucleoids. The organization of the nucleoid is a very fundamental question in mitochondrial biology and will determine tissue segregation and transmission of mtDNA. We have used a combination of stimulated emission depletion microscopy, enabling a resolution well below the diffraction barrier, and molecular biology to study nucleoids in a panel of mammalian tissue culture cells. We report that the nucleoids labeled with antibodies against DNA, mitochondrial transcription factor A (TFAM), or incorporated BrdU, have a defined, uniform mean size of ∼100 nm in mammals. Interestingly, the nucleoid frequently contains only a single copy of mtDNA (average ∼1.4 mtDNA molecules per nucleoid). Furthermore, we show by molecular modeling and volume calculations that TFAM is a main constituent of the nucleoid, besides mtDNA. These fundamental insights into the organization of mtDNA have broad implications for understanding mitochondrial dysfunction in disease and aging.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Lang BF, et al. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–497. - PubMed
-
- Nass MM. Mitochondrial DNA. I. Intramitochondrial distribution and structural relations of single- and double-length circular DNA. J Mol Biol. 1969;42:521–528. - PubMed
-
- Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. - PubMed
-
- Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26(2 Pt 2):167–180. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

