The five near-iron transporter (NEAT) domain anthrax hemophore, IsdX2, scavenges heme from hemoglobin and transfers heme to the surface protein IsdC
- PMID: 21808055
- PMCID: PMC3190864
- DOI: 10.1074/jbc.M111.241687
The five near-iron transporter (NEAT) domain anthrax hemophore, IsdX2, scavenges heme from hemoglobin and transfers heme to the surface protein IsdC
Abstract
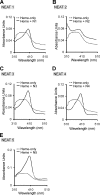
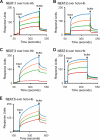
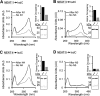
Pathogenic bacteria require iron to replicate inside mammalian hosts. Recent studies indicate that heme acquisition in Gram-positive bacteria is mediated by proteins containing one or more near-iron transporter (NEAT) domains. Bacillus anthracis is a spore-forming, Gram-positive pathogen and the causative agent of anthrax disease. The rapid, extensive, and efficient replication of B. anthracis in host tissues makes this pathogen an excellent model organism for the study of bacterial heme acquisition. B. anthracis secretes two NEAT hemophores, IsdX1 and IsdX2. IsdX1 contains a single NEAT domain, whereas IsdX2 has five, a novel property among hemophores. To understand the functional significance of harboring multiple, non-identical NEAT domains, we purified each individual NEAT domain of IsdX2 as a GST fusion and analyzed the specific function of each domain as it relates to heme acquisition and transport. NEAT domains 1, 3, 4, and 5 all bind heme, with domain 5 having the highest affinity. All NEATs associate with hemoglobin, but only NEAT1 and -5 can extract heme from hemoglobin, seemingly by a specific and active process. NEAT1, -3, and -4 transfer heme to IsdC, a cell wall-anchored anthrax NEAT protein. These results indicate that IsdX2 has all the features required to acquire heme from the host and transport heme to the bacterial cell wall. Additionally, these results suggest that IsdX2 may accelerate iron import rates by acting as a "heme sponge" that enhances B. anthracis replication in iron-starved environments.
Figures




References
-
- Crosa J. H., Mey A. R., Payne S. M. (2004) Iron Transport in Bacteria, American Society of Microbiology Press, Washington, D. C
-
- Heinemann I. U., Jahn M., Jahn D. (2008) Arch. Biochem. Biophys. 474, 238–251 - PubMed
-
- De Domenico I., McVey Ward D., Kaplan J. (2008) Nat. Rev. Mol. Cell Biol. 9, 72–81 - PubMed
-
- Wandersman C., Delepelaire P. (2004) Annu. Rev. Microbiol. 58, 611–647 - PubMed
-
- Cescau S., Cwerman H., Létoffé S., Delepelaire P., Wandersman C., Biville F. (2007) Biometals 20, 603–613 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL047020/HL/NHLBI NIH HHS/United States
- GM84348/GM/NIGMS NIH HHS/United States
- GM035649/GM/NIGMS NIH HHS/United States
- R21 AI096314/AI/NIAID NIH HHS/United States
- F32 AI069697/AI/NIAID NIH HHS/United States
- K22 AI079165/AI/NIAID NIH HHS/United States
- AI069697/AI/NIAID NIH HHS/United States
- R01 HL047020/HL/NHLBI NIH HHS/United States
- R21 AI146481/AI/NIAID NIH HHS/United States
- R21 AI088329/AI/NIAID NIH HHS/United States
- R01 GM084348/GM/NIGMS NIH HHS/United States
- R01 GM035649/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

