Pseudomonas aeruginosa Genomic Structure and Diversity
- PMID: 21808635
- PMCID: PMC3139241
- DOI: 10.3389/fmicb.2011.00150
Pseudomonas aeruginosa Genomic Structure and Diversity
Abstract
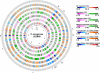
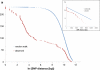
The Pseudomonas aeruginosa genome (G + C content 65-67%, size 5.5-7 Mbp) is made up of a single circular chromosome and a variable number of plasmids. Sequencing of complete genomes or blocks of the accessory genome has revealed that the genome encodes a large repertoire of transporters, transcriptional regulators, and two-component regulatory systems which reflects its metabolic diversity to utilize a broad range of nutrients. The conserved core component of the genome is largely collinear among P. aeruginosa strains and exhibits an interclonal sequence diversity of 0.5-0.7%. Only a few loci of the core genome are subject to diversifying selection. Genome diversity is mainly caused by accessory DNA elements located in 79 regions of genome plasticity that are scattered around the genome and show an anomalous usage of mono- to tetradecanucleotides. Genomic islands of the pKLC102/PAGI-2 family that integrate into tRNA(Lys) or tRNA(Gly) genes represent hotspots of inter- and intraclonal genomic diversity. The individual islands differ in their repertoire of metabolic genes that make a large contribution to the pangenome. In order to unravel intraclonal diversity of P. aeruginosa, the genomes of two members of the PA14 clonal complex from diverse habitats and geographic origin were compared. The genome sequences differed by less than 0.01% from each other. One hundred ninety-eight of the 231 single nucleotide substitutions (SNPs) were non-randomly distributed in the genome. Non-synonymous SNPs were mainly found in an integrated Pf1-like phage and in genes involved in transcriptional regulation, membrane and extracellular constituents, transport, and secretion. In summary, P. aeruginosa is endowed with a highly conserved core genome of low sequence diversity and a highly variable accessory genome that communicates with other pseudomonads and genera via horizontal gene transfer.
Keywords: Pseudomonas aeruginosa; accessory genome; clonal complex; core genome; genome; genomic island; oligonucleotide signature.
Figures




References
-
- Aendekerk S., Diggle S. P., Song Z., Høiby N., Cornelis P., Williams P., Cámara M. (2005). The MexGHI-OpmD multidrug efflux pump controls growth, antibiotic susceptibility and virulence in Pseudomonas aeruginosa via 4-quinolone-dependent cell-to-cell communication. Microbiology 151(Pt 4), 1113–112510.1099/mic.0.27631-0 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

