Insights into role of the hydrogen bond networks in substrate recognition by UDP-GalNAc 4-epimerases
- PMID: 21810411
- PMCID: PMC3441825
- DOI: 10.1016/j.bbrc.2011.07.071
Insights into role of the hydrogen bond networks in substrate recognition by UDP-GalNAc 4-epimerases
Abstract
UDP-hexose 4-epimerases are critical in galactose metabolism and often important in lipopolysaccharide biosynthesis as well. Three groups of these enzymes have been reported based on their substrate specificity towards non-acetylated substrates (group 1), dual specificity towards N-acetylated and non-acetylated substrates (group 2) and specificity towards N-acetylated substrates (group 3). We recently reported the structure of a novel UDP-GalNAc 4-epimerase called WbgU and based on the structure proposed a model of specific substrate recognition by UDP-GalNAc 4-epimerases. In this work, we present an analysis of the proposed model of substrate recognition using site-directed mutagenesis of WbgU and crystal structure of the His305Ala mutant. This investigation reveals that the wild-type activity of WbgU is retained in most single-point mutants targeting the active site. However, a graded loss in activity is observed in double-and triple-point mutants with the quadruple-point mutant being completely inactive corroborating the proposed rationale of substrate recognition. Furthermore, crystal structure of the His305Ala mutant shows that the structure is significantly similar to the wild-type WbgU, albeit a loss in the critical hydrogen bond network seated at His305 and ensuing minor conformational changes. It is inferred that the specific and non-specific interactions throughout the active site confer it sufficient elasticity to sustain wild-type activity for several of the single-point mutations.
Copyright © 2011. Published by Elsevier Inc.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

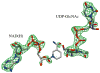
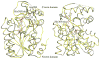
References
-
- Engh RA, Huber R. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr A. 1991;47:392–400.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

