Metabolic network reconstruction of Chlamydomonas offers insight into light-driven algal metabolism
- PMID: 21811229
- PMCID: PMC3202792
- DOI: 10.1038/msb.2011.52
Metabolic network reconstruction of Chlamydomonas offers insight into light-driven algal metabolism
Abstract
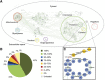
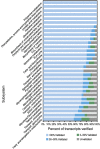
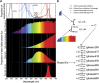
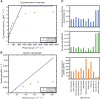
Metabolic network reconstruction encompasses existing knowledge about an organism's metabolism and genome annotation, providing a platform for omics data analysis and phenotype prediction. The model alga Chlamydomonas reinhardtii is employed to study diverse biological processes from photosynthesis to phototaxis. Recent heightened interest in this species results from an international movement to develop algal biofuels. Integrating biological and optical data, we reconstructed a genome-scale metabolic network for this alga and devised a novel light-modeling approach that enables quantitative growth prediction for a given light source, resolving wavelength and photon flux. We experimentally verified transcripts accounted for in the network and physiologically validated model function through simulation and generation of new experimental growth data, providing high confidence in network contents and predictive applications. The network offers insight into algal metabolism and potential for genetic engineering and efficient light source design, a pioneering resource for studying light-driven metabolism and quantitative systems biology.
Figures
Similar articles
-
Flux balance analysis of primary metabolism in Chlamydomonas reinhardtii.BMC Syst Biol. 2009 Jan 7;3:4. doi: 10.1186/1752-0509-3-4. BMC Syst Biol. 2009. PMID: 19128495 Free PMC article.
-
A refined genome-scale reconstruction of Chlamydomonas metabolism provides a platform for systems-level analyses.Plant J. 2015 Dec;84(6):1239-56. doi: 10.1111/tpj.13059. Epub 2015 Nov 30. Plant J. 2015. PMID: 26485611 Free PMC article.
-
AlgaGEM--a genome-scale metabolic reconstruction of algae based on the Chlamydomonas reinhardtii genome.BMC Genomics. 2011 Dec 22;12 Suppl 4(Suppl 4):S5. doi: 10.1186/1471-2164-12-S4-S5. Epub 2011 Dec 22. BMC Genomics. 2011. PMID: 22369158 Free PMC article.
-
Metabolic systems analysis to advance algal biotechnology.Biotechnol J. 2010 Jul;5(7):660-70. doi: 10.1002/biot.201000129. Biotechnol J. 2010. PMID: 20665641 Review.
-
Chlamydomonas reinhardtii as the photosynthetic yeast.Annu Rev Genet. 1995;29:209-30. doi: 10.1146/annurev.ge.29.120195.001233. Annu Rev Genet. 1995. PMID: 8825474 Review.
Cited by
-
Integration of genome-scale modeling and transcript profiling reveals metabolic pathways underlying light and temperature acclimation in Arabidopsis.Plant Cell. 2013 Apr;25(4):1197-211. doi: 10.1105/tpc.112.108852. Epub 2013 Apr 23. Plant Cell. 2013. PMID: 23613196 Free PMC article.
-
Predictive modeling of biomass component tradeoffs in Brassica napus developing oilseeds based on in silico manipulation of storage metabolism.Plant Physiol. 2012 Nov;160(3):1218-36. doi: 10.1104/pp.112.203927. Epub 2012 Sep 14. Plant Physiol. 2012. PMID: 22984123 Free PMC article.
-
Introns mediate post-transcriptional enhancement of nuclear gene expression in the green microalga Chlamydomonas reinhardtii.PLoS Genet. 2020 Jul 30;16(7):e1008944. doi: 10.1371/journal.pgen.1008944. eCollection 2020 Jul. PLoS Genet. 2020. PMID: 32730252 Free PMC article.
-
BioModTool: from biomass composition data to structured biomass objective functions for genome-scale metabolic models.Bioinform Adv. 2025 Feb 21;5(1):vbaf036. doi: 10.1093/bioadv/vbaf036. eCollection 2025. Bioinform Adv. 2025. PMID: 40065773 Free PMC article.
-
Functional genomics tools applied to plant metabolism: a survey on plant respiration, its connections and the annotation of complex gene functions.Front Plant Sci. 2012 Sep 6;3:210. doi: 10.3389/fpls.2012.00210. eCollection 2012. Front Plant Sci. 2012. PMID: 22973288 Free PMC article.
References
-
- Akkerman I, Janssen M, Rocha J, Wijffels RH (2002) Photobiological hydrogen production: photochemical efficiency and bioreactor design. Int J Hydrogen Energy 27: 1195–1208
-
- Barta DJ, Tibbitts TW, Bula RJ, Morrow RC (1992) Evaluation of light emitting diode characteristics for a space-based plant irradiation source. Adv Space Res 12: 141–149 - PubMed
-
- Becker SA, Feist AM, Mo ML, Hannum G, Palsson BO, Herrgard MJ (2007) Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat Protoc 2: 727–738 - PubMed
-
- Berg J, Tymoczko J, Stryer L (2007) Biochemistry. New York, USA: W.H. Freeman
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

