Global mRNA degradation during lytic gammaherpesvirus infection contributes to establishment of viral latency
- PMID: 21811408
- PMCID: PMC3141057
- DOI: 10.1371/journal.ppat.1002150
Global mRNA degradation during lytic gammaherpesvirus infection contributes to establishment of viral latency
Abstract
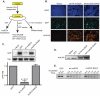
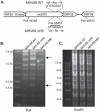
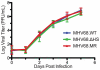
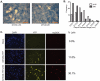
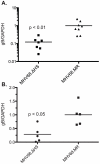
During a lytic gammaherpesvirus infection, host gene expression is severely restricted by the global degradation and altered 3' end processing of mRNA. This host shutoff phenotype is orchestrated by the viral SOX protein, yet its functional significance to the viral lifecycle has not been elucidated, in part due to the multifunctional nature of SOX. Using an unbiased mutagenesis screen of the murine gammaherpesvirus 68 (MHV68) SOX homolog, we isolated a single amino acid point mutant that is selectively defective in host shutoff activity. Incorporation of this mutation into MHV68 yielded a virus with significantly reduced capacity for mRNA turnover. Unexpectedly, the MHV68 mutant showed little defect during the acute replication phase in the mouse lung. Instead, the virus exhibited attenuation at later stages of in vivo infections suggestive of defects in both trafficking and latency establishment. Specifically, mice intranasally infected with the host shutoff mutant accumulated to lower levels at 10 days post infection in the lymph nodes, failed to develop splenomegaly, and exhibited reduced viral DNA levels and a lower frequency of latently infected splenocytes. Decreased latency establishment was also observed upon infection via the intraperitoneal route. These results highlight for the first time the importance of global mRNA degradation during a gammaherpesvirus infection and link an exclusively lytic phenomenon with downstream latency establishment.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

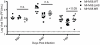

References
-
- Glaunsinger B, Ganem D. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol Cell. 2004;13:713–723. - PubMed
-
- Darnell JE, Jr, Levintow L. Poliovirus protein: source of amino acids and time course of synthesis. J Biol Chem. 1960;235:74–77. - PubMed
-
- Plotch SJ, Bouloy M, Ulmanen I, Krug RM. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847–858. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

