Mechanisms of chromosome number evolution in yeast
- PMID: 21811419
- PMCID: PMC3141009
- DOI: 10.1371/journal.pgen.1002190
Mechanisms of chromosome number evolution in yeast
Abstract
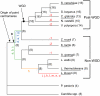
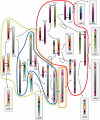
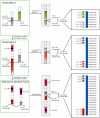
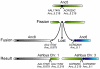
The whole-genome duplication (WGD) that occurred during yeast evolution changed the basal number of chromosomes from 8 to 16. However, the number of chromosomes in post-WGD species now ranges between 10 and 16, and the number in non-WGD species (Zygosaccharomyces, Kluyveromyces, Lachancea, and Ashbya) ranges between 6 and 8. To study the mechanism by which chromosome number changes, we traced the ancestry of centromeres and telomeres in each species. We observe only two mechanisms by which the number of chromosomes has decreased, as indicated by the loss of a centromere. The most frequent mechanism, seen 8 times, is telomere-to-telomere fusion between two chromosomes with the concomitant death of one centromere. The other mechanism, seen once, involves the breakage of a chromosome at its centromere, followed by the fusion of the two arms to the telomeres of two other chromosomes. The only mechanism by which chromosome number has increased in these species is WGD. Translocations and inversions have cycled telomere locations, internalizing some previously telomeric genes and creating novel telomeric locations. Comparison of centromere structures shows that the length of the CDEII region is variable between species but uniform within species. We trace the complete rearrangement history of the Lachancea kluyveri genome since its common ancestor with Saccharomyces and propose that its exceptionally low level of rearrangement is a consequence of the loss of the non-homologous end joining (NHEJ) DNA repair pathway in this species.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Spatial inter-centromeric interactions facilitated the emergence of evolutionary new centromeres.Elife. 2020 May 29;9:e58556. doi: 10.7554/eLife.58556. Elife. 2020. PMID: 32469306 Free PMC article.
-
Chromosome number reduction in Eremothecium coryli by two telomere-to-telomere fusions.Genome Biol Evol. 2014 May 6;6(5):1186-98. doi: 10.1093/gbe/evu089. Genome Biol Evol. 2014. PMID: 24803574 Free PMC article.
-
Chromosomal G + C content evolution in yeasts: systematic interspecies differences, and GC-poor troughs at centromeres.Genome Biol Evol. 2010;2:572-83. doi: 10.1093/gbe/evq042. Epub 2010 Jul 8. Genome Biol Evol. 2010. PMID: 20693156 Free PMC article.
-
Yeast genome evolution--the origin of the species.Yeast. 2007 Nov;24(11):929-42. doi: 10.1002/yea.1515. Yeast. 2007. PMID: 17621376 Review.
-
Telomeres and chromosome instability.DNA Repair (Amst). 2006 Sep 8;5(9-10):1082-92. doi: 10.1016/j.dnarep.2006.05.030. Epub 2006 Jun 19. DNA Repair (Amst). 2006. PMID: 16784900 Review.
Cited by
-
Loss of inner kinetochore genes is associated with the transition to an unconventional point centromere in budding yeast.PeerJ. 2020 Sep 29;8:e10085. doi: 10.7717/peerj.10085. eCollection 2020. PeerJ. 2020. PMID: 33062452 Free PMC article.
-
A molecular genetic toolbox for Yarrowia lipolytica.Biotechnol Biofuels. 2017 Jan 3;10:2. doi: 10.1186/s13068-016-0687-7. eCollection 2017. Biotechnol Biofuels. 2017. PMID: 28066508 Free PMC article.
-
Diversity of mating-type chromosome structures in the yeast Zygosaccharomyces rouxii caused by ectopic exchanges between MAT-like loci.PLoS One. 2013 Apr 16;8(4):e62121. doi: 10.1371/journal.pone.0062121. Print 2013. PLoS One. 2013. PMID: 23614024 Free PMC article.
-
Experimental Evolution Reveals Interplay between Sch9 and Polyploid Stability in Yeast.PLoS Genet. 2016 Nov 3;12(11):e1006409. doi: 10.1371/journal.pgen.1006409. eCollection 2016 Nov. PLoS Genet. 2016. PMID: 27812096 Free PMC article.
-
Polymorphic centromere locations in the pathogenic yeast Candida parapsilosis.Genome Res. 2020 May;30(5):684-696. doi: 10.1101/gr.257816.119. Epub 2020 May 18. Genome Res. 2020. PMID: 32424070 Free PMC article.
References
-
- Hillier LW, Graves TA, Fulton RS, Fulton LA, Pepin KH, et al. Generation and annotation of the DNA sequences of human chromosomes 2 and 4. Nature. 2005;434:724–731. - PubMed
-
- International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

