SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV
- PMID: 21811420
- PMCID: PMC3141008
- DOI: 10.1371/journal.pgen.1002195
SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV
Abstract
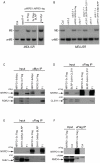
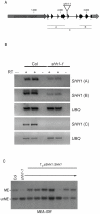
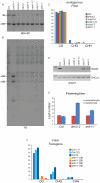
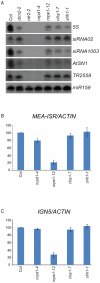
DNA methylation is an evolutionarily conserved epigenetic modification that is critical for gene silencing and the maintenance of genome integrity. In Arabidopsis thaliana, the de novo DNA methyltransferase, domains rearranged methyltransferase 2 (DRM2), is targeted to specific genomic loci by 24 nt small interfering RNAs (siRNAs) through a pathway termed RNA-directed DNA methylation (RdDM). Biogenesis of the targeting siRNAs is thought to be initiated by the activity of the plant-specific RNA polymerase IV (Pol-IV). However, the mechanism through which Pol-IV is targeted to specific genomic loci and whether factors other than the core Pol-IV machinery are required for Pol-IV activity remain unknown. Through the affinity purification of nuclear RNA polymerase D1 (NRPD1), the largest subunit of the Pol-IV polymerase, we found that several previously identified RdDM components co-purify with Pol-IV, namely RNA-dependent RNA polymerase 2 (RDR2), CLASSY1 (CLSY1), and RNA-directed DNA methylation 4 (RDM4), suggesting that the upstream siRNA generating portion of the RdDM pathway may be more physically coupled than previously envisioned. A homeodomain protein, SAWADEE homeodomain homolog 1 (SHH1), was also found to co-purify with NRPD1; and we demonstrate that SHH1 is required for de novo and maintenance DNA methylation, as well as for the accumulation of siRNAs at specific loci, confirming it is a bonafide component of the RdDM pathway.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1.Nature. 2013 Jun 20;498(7454):385-9. doi: 10.1038/nature12178. Epub 2013 May 1. Nature. 2013. PMID: 23636332 Free PMC article.
-
DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV.Proc Natl Acad Sci U S A. 2013 May 14;110(20):8290-5. doi: 10.1073/pnas.1300585110. Epub 2013 May 1. Proc Natl Acad Sci U S A. 2013. PMID: 23637343 Free PMC article.
-
The Pol IV largest subunit CTD quantitatively affects siRNA levels guiding RNA-directed DNA methylation.Nucleic Acids Res. 2019 Sep 26;47(17):9024-9036. doi: 10.1093/nar/gkz615. Nucleic Acids Res. 2019. PMID: 31329950 Free PMC article.
-
Advances of RNA polymerase IV in controlling DNA methylation and development in plants.Yi Chuan. 2022 Jul 20;44(7):567-580. doi: 10.16288/j.yczz.22-063. Yi Chuan. 2022. PMID: 35858769 Review.
-
RNA-directed DNA methylation in plants: Where to start?RNA Biol. 2013 Oct;10(10):1593-6. doi: 10.4161/rna.26312. RNA Biol. 2013. PMID: 25003825 Free PMC article. Review.
Cited by
-
ZMP recruits and excludes Pol IV-mediated DNA methylation in a site-specific manner.Sci Adv. 2022 Nov 25;8(47):eadc9454. doi: 10.1126/sciadv.adc9454. Epub 2022 Nov 25. Sci Adv. 2022. PMID: 36427317 Free PMC article.
-
Multifaceted roles of RNA polymerase IV in plant growth and development.J Exp Bot. 2020 Oct 7;71(19):5725-5732. doi: 10.1093/jxb/eraa346. J Exp Bot. 2020. PMID: 32969476 Free PMC article.
-
Identification of transcription factor binding sites on promoter of RNA dependent RNA polymerases (RDRs) and interacting partners of RDR proteins through in silico analysis.Physiol Mol Biol Plants. 2019 Jul;25(4):1055-1071. doi: 10.1007/s12298-019-00660-w. Epub 2019 May 6. Physiol Mol Biol Plants. 2019. PMID: 31402824 Free PMC article.
-
Detailed insight into the dynamics of the initial phases of de novo RNA-directed DNA methylation in plant cells.Epigenetics Chromatin. 2019 Sep 11;12(1):54. doi: 10.1186/s13072-019-0299-0. Epigenetics Chromatin. 2019. PMID: 31511048 Free PMC article.
-
Evidence for a Unique DNA-Dependent RNA Polymerase in Cereal Crops.Mol Biol Evol. 2018 Oct 1;35(10):2454-2462. doi: 10.1093/molbev/msy146. Mol Biol Evol. 2018. PMID: 30053133 Free PMC article.
References
-
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. - PubMed
-
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. - PubMed
-
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–376. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

