Protein Secretion Systems in Pseudomonas aeruginosa: An Essay on Diversity, Evolution, and Function
- PMID: 21811488
- PMCID: PMC3140646
- DOI: 10.3389/fmicb.2011.00155
Protein Secretion Systems in Pseudomonas aeruginosa: An Essay on Diversity, Evolution, and Function
Abstract
Protein secretion systems are molecular nanomachines used by Gram-negative bacteria to thrive within their environment. They are used to release enzymes that hydrolyze complex carbon sources into usable compounds, or to release proteins that capture essential ions such as iron. They are also used to colonize and survive within eukaryotic hosts, causing acute or chronic infections, subverting the host cell response and escaping the immune system. In this article, the opportunistic human pathogen Pseudomonas aeruginosa is used as a model to review the diversity of secretion systems that bacteria have evolved to achieve these goals. This diversity may result from a progressive transformation of cell envelope complexes that initially may not have been dedicated to secretion. The striking similarities between secretion systems and type IV pili, flagella, bacteriophage tail, or efflux pumps is a nice illustration of this evolution. Differences are also needed since various secretion configurations call for diversity. For example, some proteins are released in the extracellular medium while others are directly injected into the cytosol of eukaryotic cells. Some proteins are folded before being released and transit into the periplasm. Other proteins cross the whole cell envelope at once in an unfolded state. However, the secretion system requires conserved basic elements or features. For example, there is a need for an energy source or for an outer membrane channel. The structure of this review is thus quite unconventional. Instead of listing secretion types one after each other, it presents a melting pot of concepts indicating that secretion types are in constant evolution and use basic principles. In other words, emergence of new secretion systems could be predicted the way Mendeleïev had anticipated characteristics of yet unknown elements.
Keywords: cell envelope; channel; macromolecular complex; nanomachine; targeting.
Figures
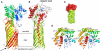

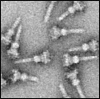


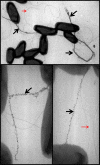

Similar articles
-
Structural Basis of Type 2 Secretion System Engagement between the Inner and Outer Bacterial Membranes.mBio. 2017 Oct 17;8(5):e01344-17. doi: 10.1128/mBio.01344-17. mBio. 2017. PMID: 29042496 Free PMC article.
-
Pseudomonas aeruginosa Pore-Forming Exolysin and Type IV Pili Cooperate To Induce Host Cell Lysis.mBio. 2017 Jan 24;8(1):e02250-16. doi: 10.1128/mBio.02250-16. mBio. 2017. PMID: 28119472 Free PMC article.
-
The HsiB1C1 (TssB-TssC) complex of the Pseudomonas aeruginosa type VI secretion system forms a bacteriophage tail sheathlike structure.J Biol Chem. 2013 Mar 15;288(11):7536-7548. doi: 10.1074/jbc.M112.439273. Epub 2013 Jan 22. J Biol Chem. 2013. PMID: 23341461 Free PMC article.
-
Protein secretion systems in Pseudomonas aeruginosa: A wealth of pathogenic weapons.Int J Med Microbiol. 2010 Dec;300(8):534-43. doi: 10.1016/j.ijmm.2010.08.005. Epub 2010 Oct 13. Int J Med Microbiol. 2010. PMID: 20947426 Review.
-
Channel-tunnels: outer membrane components of type I secretion systems and multidrug efflux pumps of Gram-negative bacteria.Rev Physiol Biochem Pharmacol. 2003;147:122-65. doi: 10.1007/s10254-003-0008-y. Epub 2003 Mar 13. Rev Physiol Biochem Pharmacol. 2003. PMID: 12783268 Review.
Cited by
-
A Review of the Functional Annotations of Important Genes in the AHPND-Causing pVA1 Plasmid.Microorganisms. 2020 Jul 3;8(7):996. doi: 10.3390/microorganisms8070996. Microorganisms. 2020. PMID: 32635298 Free PMC article. Review.
-
TypA is involved in virulence, antimicrobial resistance and biofilm formation in Pseudomonas aeruginosa.BMC Microbiol. 2013 Apr 9;13:77. doi: 10.1186/1471-2180-13-77. BMC Microbiol. 2013. PMID: 23570569 Free PMC article.
-
Global control of GacA in secondary metabolism, primary metabolism, secretion systems, and motility in the rhizobacterium Pseudomonas aeruginosa M18.J Bacteriol. 2013 Aug;195(15):3387-400. doi: 10.1128/JB.00214-13. Epub 2013 May 24. J Bacteriol. 2013. PMID: 23708134 Free PMC article.
-
Pseudomonas aeruginosa Triggered Exosomal Release of ADAM10 Mediates Proteolytic Cleavage in Trans.Int J Mol Sci. 2022 Jan 23;23(3):1259. doi: 10.3390/ijms23031259. Int J Mol Sci. 2022. PMID: 35163191 Free PMC article.
-
EEF2-inactivating toxins engage the NLRP1 inflammasome and promote epithelial barrier disruption.J Exp Med. 2023 Oct 2;220(10):e20230104. doi: 10.1084/jem.20230104. Epub 2023 Aug 29. J Exp Med. 2023. PMID: 37642996 Free PMC article.
References
-
- Akrim M., Bally M., Ball G., Tommassen J., Teerink H., Filloux A., Lazdunski A. (1993). Xcp-mediated protein secretion in Pseudomonas aeruginosa: identification of two additional genes and evidence for regulation of xcp gene expression. Mol. Microbiol. 10, 431–443 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

