Emergence of a globally dominant IncHI1 plasmid type associated with multiple drug resistant typhoid
- PMID: 21811646
- PMCID: PMC3139670
- DOI: 10.1371/journal.pntd.0001245
Emergence of a globally dominant IncHI1 plasmid type associated with multiple drug resistant typhoid
Abstract
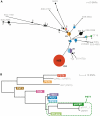
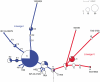
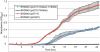
Typhoid fever, caused by Salmonella enterica serovar Typhi (S. Typhi), remains a serious global health concern. Since their emergence in the mid-1970s multi-drug resistant (MDR) S. Typhi now dominate drug sensitive equivalents in many regions. MDR in S. Typhi is almost exclusively conferred by self-transmissible IncHI1 plasmids carrying a suite of antimicrobial resistance genes. We identified over 300 single nucleotide polymorphisms (SNPs) within conserved regions of the IncHI1 plasmid, and genotyped both plasmid and chromosomal SNPs in over 450 S. Typhi dating back to 1958. Prior to 1995, a variety of IncHI1 plasmid types were detected in distinct S. Typhi haplotypes. Highly similar plasmids were detected in co-circulating S. Typhi haplotypes, indicative of plasmid transfer. In contrast, from 1995 onwards, 98% of MDR S. Typhi were plasmid sequence type 6 (PST6) and S. Typhi haplotype H58, indicating recent global spread of a dominant MDR clone. To investigate whether PST6 conferred a selective advantage compared to other IncHI1 plasmids, we used a phenotyping array to compare the impact of IncHI1 PST6 and PST1 plasmids in a common S. Typhi host. The PST6 plasmid conferred the ability to grow in high salt medium (4.7% NaCl), which we demonstrate is due to the presence in PST6 of the Tn6062 transposon encoding BetU.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Kothari A, Pruthi A, Chugh TD. The burden of enteric fever. J Infect Dev Ctries. 2008;2:253–259. - PubMed
-
- World Health Organization. Communicable Disease Surveillance and Response Vaccine and Biologicals; 2003. The diagnosis, treatment and prevention of typhoid fever. pp. 7–18.
-
- Whitaker JA, Franco-Paredes C, del Rio C, Edupuganti S. Rethinking typhoid fever vaccines: implications for travelers and people living in highly endemic areas. J Travel Med. 2009;16:46–52. - PubMed
-
- Ochiai RL, Acosta CJ, Agtini M, Bhattacharya SK, Bhutta ZA, et al. The use of typhoid vaccines in Asia: the DOMI experience. Clin Infect Dis. 2007;45(Suppl 1):S34–38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

