A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin
- PMID: 21813488
- PMCID: PMC3175761
- DOI: 10.1093/gbe/evr078
A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin
Abstract
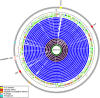
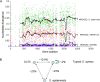
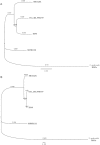
Here we discuss the evolution of the northern Australian Staphylococcus aureus isolate MSHR1132 genome. MSHR1132 belongs to the divergent clonal complex 75 lineage. The average nucleotide divergence between orthologous genes in MSHR1132 and typical S. aureus is approximately sevenfold greater than the maximum divergence observed in this species to date. MSHR1132 has a small accessory genome, which includes the well-characterized genomic islands, νSAα and νSaβ, suggesting that these elements were acquired well before the expansion of the typical S. aureus population. Other mobile elements show mosaic structure (the prophage ϕSa3) or evidence of recent acquisition from a typical S. aureus lineage (SCCmec, ICE6013 and plasmid pMSHR1132). There are two differences in gene repertoire compared with typical S. aureus that may be significant clues as to the genetic basis underlying the successful emergence of S. aureus as a pathogen. First, MSHR1132 lacks the genes for production of staphyloxanthin, the carotenoid pigment that confers upon S. aureus its characteristic golden color and protects against oxidative stress. The lack of pigment was demonstrated in 126 of 126 CC75 isolates. Second, a mobile clustered regularly interspaced short palindromic repeat (CRISPR) element is inserted into orfX of MSHR1132. Although common in other staphylococcal species, these elements are very rare within S. aureus and may impact accessory genome acquisition. The CRISPR spacer sequences reveal a history of attempted invasion by known S. aureus mobile elements. There is a case for the creation of a new taxon to accommodate this and related isolates.
Figures




References
-
- Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol. 2008;6:431–440. - PubMed
-
- Altschul SF, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Baba T, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

