HLA-DR+ CD38+ CD4+ T lymphocytes have elevated CCR5 expression and produce the majority of R5-tropic HIV-1 RNA in vivo
- PMID: 21813616
- PMCID: PMC3196402
- DOI: 10.1128/JVI.02529-10
HLA-DR+ CD38+ CD4+ T lymphocytes have elevated CCR5 expression and produce the majority of R5-tropic HIV-1 RNA in vivo
Abstract
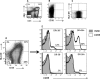
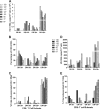
Percentages of activated T cells correlate with HIV-1 disease progression, but the underlying mechanisms are not fully understood. We hypothesized that HLA-DR(+) CD38(+) (DR(+) 38(+)) CD4(+) T cells produce the majority of HIV-1 due to elevated expression of CCR5 and CXCR4. In phytohemagglutinin (PHA)-stimulated CD8-depleted peripheral blood mononuclear cells (PBMC) infected with HIV-1 green fluorescent protein (GFP) reporter viruses, DR(-) 38(+) T cells constituted the majority of CCR5 (R5)-tropic (median, 62%) and CXCR4 (X4)-tropic HIV-1-producing cells (median, 61%), although cell surface CCR5 and CXCR4 were not elevated in this subset of cells. In lymph nodes from untreated individuals infected with R5-tropic HIV-1, percentages of CCR5(+) cells were elevated in DR(+) 38(+) CD4(+) T cells (median, 36.4%) compared to other CD4(+) T-cell subsets (median values of 5.7% for DR(-) 38(-) cells, 19.4% for DR(+) 38(-) cells, and 7.6% for DR(-) 38(+) cells; n = 18; P < 0.001). In sorted CD8(-) lymph node T cells, median HIV-1 RNA copies/10(5) cells was higher for DR(+) 38(+) cells (1.8 × 10(6)) than for DR(-) 38(-) (0.007 × 10(6)), DR(-) 38(+) (0.064 × 10(6)), and DR(+) 38(-) (0.18 × 10(6)) subsets (n = 8; P < 0.001 for all). After adjusting for percentages of subsets, a median of 87% of viral RNA was harbored by DR(+) 38(+) cells. Percentages of CCR5(+) CD4(+) T cells and concentrations of CCR5 molecules among subsets predicted HIV-1 RNA levels among CD8(-) DR/38 subsets (P < 0.001 for both). Median HIV-1 DNA copies/10(5) cells was higher in DR(+) 38(+) cells (5,360) than in the DR(-) 38(-) (906), DR(-) 38(+) (814), and DR(+) 38(-) (1,984) subsets (n = 7; P ≤ 0.031). Thus, DR(+) 38(+) CD4(+) T cells in lymph nodes have elevated CCR5 expression, are highly susceptible to infection with R5-tropic virus, and produce the majority of R5-tropic HIV-1. PBMC assays failed to recapitulate in vivo findings, suggesting limited utility. Strategies to reduce numbers of DR(+) 38(+) CD4(+) T cells may substantially inhibit HIV-1 replication.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

