The tomato terpene synthase gene family
- PMID: 21813655
- PMCID: PMC3192577
- DOI: 10.1104/pp.111.179648
The tomato terpene synthase gene family
Abstract
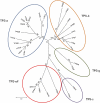
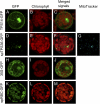
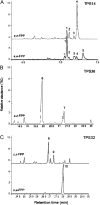
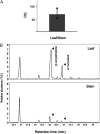
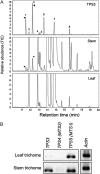
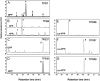
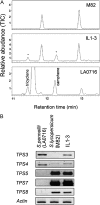
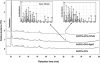
Compounds of the terpenoid class play numerous roles in the interactions of plants with their environment, such as attracting pollinators and defending the plant against pests. We show here that the genome of cultivated tomato (Solanum lycopersicum) contains 44 terpene synthase (TPS) genes, including 29 that are functional or potentially functional. Of these 29 TPS genes, 26 were expressed in at least some organs or tissues of the plant. The enzymatic functions of eight of the TPS proteins were previously reported, and here we report the specific in vitro catalytic activity of 10 additional tomato terpene synthases. Many of the tomato TPS genes are found in clusters, notably on chromosomes 1, 2, 6, 8, and 10. All TPS family clades previously identified in angiosperms are also present in tomato. The largest clade of functional TPS genes found in tomato, with 12 members, is the TPS-a clade, and it appears to encode only sesquiterpene synthases, one of which is localized to the mitochondria, while the rest are likely cytosolic. A few additional sesquiterpene synthases are encoded by TPS-b clade genes. Some of the tomato sesquiterpene synthases use z,z-farnesyl diphosphate in vitro as well, or more efficiently than, the e,e-farnesyl diphosphate substrate. Genes encoding monoterpene synthases are also prevalent, and they fall into three clades: TPS-b, TPS-g, and TPS-e/f. With the exception of two enzymes involved in the synthesis of ent-kaurene, the precursor of gibberellins, no other tomato TPS genes could be demonstrated to encode diterpene synthases so far.
Figures



References
-
- Ament K, Van Schie CC, Bouwmeester HJ, Haring MA, Schuurink RC. (2006) Induction of a leaf specific geranylgeranyl pyrophosphate synthase and emission of (E,E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene in tomato are dependent on both jasmonic acid and salicylic acid signaling pathways. Planta 224: 1197–1208 - PubMed
-
- Arimura G, Huber DP, Bohlmann J. (2004) Forest tent caterpillars (Malacosoma disstria) induce local and systemic diurnal emissions of terpenoid volatiles in hybrid poplar (Populus trichocarpa x deltoides): cDNA cloning, functional characterization, and patterns of gene expression of (−)-germacrene D synthase, PtdTPS1. Plant J 37: 603–616 - PubMed
-
- Aubourg S, Lecharny A, Bohlmann J. (2002) Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Genet Genomics 267: 730–745 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

