A novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans
- PMID: 21814493
- PMCID: PMC3144186
- DOI: 10.1371/journal.pbio.1001115
A novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans
Abstract
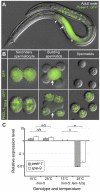
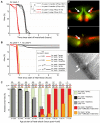
The evolutionary fate of an allele ordinarily depends on its contribution to host fitness. Occasionally, however, genetic elements arise that are able to gain a transmission advantage while simultaneously imposing a fitness cost on their hosts. We previously discovered one such element in C. elegans that gains a transmission advantage through a combination of paternal-effect killing and zygotic self-rescue. Here we demonstrate that this element is composed of a sperm-delivered toxin, peel-1, and an embryo-expressed antidote, zeel-1. peel-1 and zeel-1 are located adjacent to one another in the genome and co-occur in an insertion/deletion polymorphism. peel-1 encodes a novel four-pass transmembrane protein that is expressed in sperm and delivered to the embryo via specialized, sperm-specific vesicles. In the absence of zeel-1, sperm-delivered PEEL-1 causes lethal defects in muscle and epidermal tissue at the 2-fold stage of embryogenesis. zeel-1 is expressed transiently in the embryo and encodes a novel six-pass transmembrane domain fused to a domain with sequence similarity to zyg-11, a substrate-recognition subunit of an E3 ubiquitin ligase. zeel-1 appears to have arisen recently, during an expansion of the zyg-11 family, and the transmembrane domain of zeel-1 is required and partially sufficient for antidote activity. Although PEEL-1 and ZEEL-1 normally function in embryos, these proteins can act at other stages as well. When expressed ectopically in adults, PEEL-1 kills a variety of cell types, and ectopic expression of ZEEL-1 rescues these effects. Our results demonstrate that the tight physical linkage between two novel transmembrane proteins has facilitated their co-evolution into an element capable of promoting its own transmission to the detriment of organisms carrying it.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Hurst G. D. D, Werren J. H. The role of selfish genetic elements in eukaryotic evolution. Nature Reviews Genetics. 2001;2:597–606. - PubMed
-
- Dawe R. K, Hiatt E. N. Plant neocentromeres: fast, focused, and driven. Chromosome Res. 2004;12:655–669. - PubMed
-
- Pardo-Manuel de Villena F, Sapienza C. Nonrandom segregation during meiosis: the unfairness of females. Mamm Genome. 2001;12:331–339. - PubMed
-
- Jaenike J. Sex chromosome meiotic drive. Annual Review of Ecology and Systematics. 2001;32:25–49.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

