High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment
- PMID: 21814577
- PMCID: PMC3141039
- DOI: 10.1371/journal.pone.0022348
High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment
Abstract
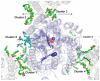
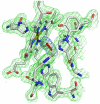
The fundamental chemistry underpinning aerobic life on Earth involves reduction of dioxygen to water with concomitant proton translocation. This process is catalyzed by members of the heme-copper oxidase (HCO) superfamily. Despite the availability of crystal structures for all types of HCO, the mode of action for this enzyme is not understood at the atomic level, namely how vectorial H(+) and e(-) transport are coupled. Toward addressing this problem, we report wild type and A120F mutant structures of the ba(3)-type cytochrome c oxidase from Thermus thermophilus at 1.8 Å resolution. The enzyme has been crystallized from the lipidic cubic phase, which mimics the biological membrane environment. The structures reveal 20 ordered lipid molecules that occupy binding sites on the protein surface or mediate crystal packing interfaces. The interior of the protein encloses 53 water molecules, including 3 trapped in the designated K-path of proton transfer and 8 in a cluster seen also in A-type enzymes that likely functions in egress of product water and proton translocation. The hydrophobic O(2)-uptake channel, connecting the active site to the lipid bilayer, contains a single water molecule nearest the Cu(B) atom but otherwise exhibits no residual electron density. The active site contains strong electron density for a pair of bonded atoms bridging the heme Fe(a3) and Cu(B) atoms that is best modeled as peroxide. The structure of ba(3)-oxidase reveals new information about the positioning of the enzyme within the membrane and the nature of its interactions with lipid molecules. The atomic resolution details provide insight into the mechanisms of electron transfer, oxygen diffusion into the active site, reduction of oxygen to water, and pumping of protons across the membrane. The development of a robust system for production of ba(3)-oxidase crystals diffracting to high resolution, together with an established expression system for generating mutants, opens the door for systematic structure-function studies.
Conflict of interest statement
Figures
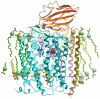


Similar articles
-
Crystallographic studies of Xe and Kr binding within the large internal cavity of cytochrome ba3 from Thermus thermophilus: structural analysis and role of oxygen transport channels in the heme-Cu oxidases.Biochemistry. 2008 Apr 22;47(16):4657-65. doi: 10.1021/bi800045y. Epub 2008 Apr 1. Biochemistry. 2008. PMID: 18376849
-
Discrete Ligand Binding and Electron Transfer Properties of ba3-Cytochrome c Oxidase from Thermus thermophilus: Evolutionary Adaption to Low Oxygen and High Temperature Environments.Acc Chem Res. 2019 May 21;52(5):1380-1390. doi: 10.1021/acs.accounts.9b00052. Epub 2019 Apr 25. Acc Chem Res. 2019. PMID: 31021078
-
Toward a chemical mechanism of proton pumping by the B-type cytochrome c oxidases: application of density functional theory to cytochrome ba3 of Thermus thermophilus.J Am Chem Soc. 2008 Nov 12;130(45):15002-21. doi: 10.1021/ja803112w. Epub 2008 Oct 17. J Am Chem Soc. 2008. PMID: 18928258 Free PMC article.
-
Proton transfer in ba(3) cytochrome c oxidase from Thermus thermophilus.Biochim Biophys Acta. 2012 Apr;1817(4):650-7. doi: 10.1016/j.bbabio.2011.11.015. Epub 2011 Dec 7. Biochim Biophys Acta. 2012. PMID: 22172736 Review.
-
Bioenergetics at extreme temperature: Thermus thermophilus ba(3)- and caa(3)-type cytochrome c oxidases.Biochim Biophys Acta. 2012 Apr;1817(4):638-49. doi: 10.1016/j.bbabio.2011.08.004. Biochim Biophys Acta. 2012. PMID: 22385645 Review.
Cited by
-
Structure of the cytochrome aa3 -600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.Proc Natl Acad Sci U S A. 2020 Jan 14;117(2):872-876. doi: 10.1073/pnas.1915013117. Epub 2019 Dec 30. Proc Natl Acad Sci U S A. 2020. PMID: 31888984 Free PMC article.
-
DFT Calculations for Mössbauer Properties on Dinuclear Center Models of the Resting Oxidized Cytochrome c Oxidase.Chemphyschem. 2022 Apr 5;23(7):e202100831. doi: 10.1002/cphc.202100831. Epub 2022 Mar 1. Chemphyschem. 2022. PMID: 35142420 Free PMC article.
-
A Water Dimer Shift Activates a Proton Pumping Pathway in the PR → F Transition of ba3 Cytochrome c Oxidase.Inorg Chem. 2018 Feb 5;57(3):1048-1059. doi: 10.1021/acs.inorgchem.7b02461. Epub 2018 Jan 8. Inorg Chem. 2018. PMID: 29308889 Free PMC article.
-
Bacterial denitrifying nitric oxide reductases and aerobic respiratory terminal oxidases use similar delivery pathways for their molecular substrates.Biochim Biophys Acta Bioenerg. 2018 Sep;1859(9):712-724. doi: 10.1016/j.bbabio.2018.06.002. Epub 2018 Jun 5. Biochim Biophys Acta Bioenerg. 2018. PMID: 29883591 Free PMC article.
-
A 2.2 Å cryoEM structure of a quinol-dependent NO Reductase shows close similarity to respiratory oxidases.Nat Commun. 2023 Jun 9;14(1):3416. doi: 10.1038/s41467-023-39140-x. Nat Commun. 2023. PMID: 37296134 Free PMC article.
References
-
- Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. - PubMed
-
- Yoshikawa S, Shinzawa-Itoh K, Nakashima R, Yaono R, Yamashita E, et al. Redox-coupled crystal structural changes in bovine heart cytochrome c oxidase. Science. 1998;280:1723–1729. - PubMed
-
- Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. - PubMed
-
- Koepke J, Olkhova E, Angerer H, Muller H, Peng G, et al. High resolution crystal structure of Paracoccus denitrificans cytochrome c oxidase: new insights into the active site and the proton transfer pathways. Biochim Biophys Acta. 2009;1787:635–645. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

