Identification of the receptor tyrosine kinase AXL in breast cancer as a target for the human miR-34a microRNA
- PMID: 21814748
- PMCID: PMC3381742
- DOI: 10.1007/s10549-011-1690-0
Identification of the receptor tyrosine kinase AXL in breast cancer as a target for the human miR-34a microRNA
Abstract
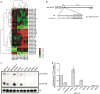
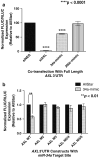
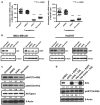
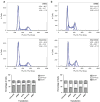
The identification of molecular features that contribute to the progression of breast cancer can provide valuable insight into the pathogenesis of this disease. Deregulated microRNA expression represents one type of molecular event that has been associated with many different human cancers. In order to identify a miRNA/mRNA regulatory interaction that is biologically relevant to the triple-negative breast cancer genotype/phenotype, we initially conducted a miRNA profiling experiment to detect differentially expressed miRNAs in cell line models representing triple-negative (MDA-MB-231), ER(+) (MCF7), and HER-2 overexpressed (SK-BR-3) histotypes. We identified human miR-34a expression as being >3-fold down (from its median expression value across all cell lines) in MDA-MB-231 cells, and identified AXL as a putative mRNA target using multiple miRNA/target prediction algorithms. The miR-34a/AXL interaction was functionally characterized through ectopic overexpression experiments with a miR-34a mimic in two independent triple-negative breast cancer cell lines. In reporter assays, miR-34a binds to its putative target site within the AXL 3'UTR to inhibit luciferase expression. We also observed degradation of AXL mRNA and decreased AXL protein levels, as well as cell signaling effects on AKT phosphorylation and phenotypic effects on cell migration. Finally, we present an inverse correlative trend in miR-34a and AXL expression for both cell line and patient tumor samples.
Conflict of interest statement
Figures


References
-
- Rakha EA, Ellis IO. Triple-negative/basal-like breast cancer: review. Pathology. 2009;41:40–47. - PubMed
-
- Venkitaraman R. Triple-negative/basal-like breast cancer: clinical, pathologic and molecular features. Expert Rev Anti-cancer Ther. 2010;10:199–207. - PubMed
-
- Charafe-Jauffret E, Ginestier C, Monville F, Finetti P, Adélaïde J, Cervera N, Fekairi S, Xerri L, Jacquemier J, Birnbaum D, Bertucci F. Gene expression profiling of breast cancer cell lines identifies potential new basal markers. Oncogene. 2006;25:2273–2284. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

