Cruciform structures are a common DNA feature important for regulating biological processes
- PMID: 21816114
- PMCID: PMC3176155
- DOI: 10.1186/1471-2199-12-33
Cruciform structures are a common DNA feature important for regulating biological processes
Abstract
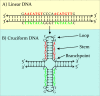
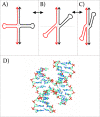
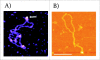
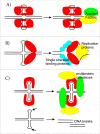
DNA cruciforms play an important role in the regulation of natural processes involving DNA. These structures are formed by inverted repeats, and their stability is enhanced by DNA supercoiling. Cruciform structures are fundamentally important for a wide range of biological processes, including replication, regulation of gene expression, nucleosome structure and recombination. They also have been implicated in the evolution and development of diseases including cancer, Werner's syndrome and others.Cruciform structures are targets for many architectural and regulatory proteins, such as histones H1 and H5, topoisomerase IIβ, HMG proteins, HU, p53, the proto-oncogene protein DEK and others. A number of DNA-binding proteins, such as the HMGB-box family members, Rad54, BRCA1 protein, as well as PARP-1 polymerase, possess weak sequence specific DNA binding yet bind preferentially to cruciform structures. Some of these proteins are, in fact, capable of inducing the formation of cruciform structures upon DNA binding. In this article, we review the protein families that are involved in interacting with and regulating cruciform structures, including (a) the junction-resolving enzymes, (b) DNA repair proteins and transcription factors, (c) proteins involved in replication and (d) chromatin-associated proteins. The prevalence of cruciform structures and their roles in protein interactions, epigenetic regulation and the maintenance of cell homeostasis are also discussed.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

