BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages
- PMID: 21816365
- PMCID: PMC3567433
- DOI: 10.1016/j.stem.2011.06.015
BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages
Abstract
BMP is thought to induce hESC differentiation toward multiple lineages including mesoderm and trophoblast. The BMP-induced trophoblast phenotype is a long-standing paradox in stem cell biology. Here we readdressed BMP function in hESCs and mouse epiblast-derived cells. We found that BMP4 cooperates with FGF2 (via ERK) to induce mesoderm and to inhibit endoderm differentiation. These conditions induced cells with high levels of BRACHYURY (BRA) that coexpressed CDX2. BRA was necessary for and preceded CDX2 expression; both genes were essential for expression not only of mesodermal genes but also of trophoblast-associated genes. Maximal expression of the latter was seen in the absence of FGF but these cells coexpressed mesodermal genes and moreover they differed in cell surface and epigenetic properties from placental trophoblast. We conclude that BMP induces human and mouse pluripotent stem cells primarily to form mesoderm, rather than trophoblast, acting through BRA and CDX2.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

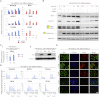



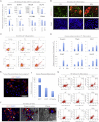
Comment in
-
When BMP meets FGF.Cell Stem Cell. 2011 Aug 5;9(2):91-2. doi: 10.1016/j.stem.2011.07.004. Cell Stem Cell. 2011. PMID: 21816358
References
-
- Apps R., Murphy S.P., Fernando R., Gardner L., Ahad T., Moffett A. Human leucocyte antigen (HLA) expression of primary trophoblast cells and placental cell lines, determined using single antigen beads to characterize allotype specificities of anti-HLA antibodies. Immunology. 2009;127:26–39. - PMC - PubMed
-
- Beck F., Erler T., Russell A., James R. Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev. Dyn. 1995;204:219–227. - PubMed
-
- Beddington R.S., Robertson E.J. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development. 1989;105:733–737. - PubMed
-
- Beddington R.S., Rashbass P., Wilson V. Brachyury—a gene affecting mouse gastrulation and early organogenesis. Dev. Suppl. 1992:157–165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

