Crohn's disease and genetic hitchhiking at IBD5
- PMID: 21816865
- PMCID: PMC3245542
- DOI: 10.1093/molbev/msr151
Crohn's disease and genetic hitchhiking at IBD5
Abstract
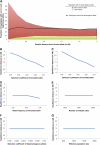
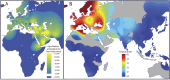
Inflammatory bowel disease 5 (IBD5) is a 250 kb haplotype on chromosome 5 that is associated with an increased risk of Crohn's disease in Europeans. The OCTN1 gene is centrally located on IBD5 and encodes a transporter of the antioxidant ergothioneine (ET). The 503F variant of OCTN1 is strongly associated with IBD5 and is a gain-of-function mutation that increases absorption of ET. Although 503F has been implicated as the variant potentially responsible for Crohn's disease susceptibility at IBD5, there is little evidence beyond statistical association to support its role in disease causation. We hypothesize that 503F is a recent adaptation in Europeans that swept to relatively high frequency and that disease association at IBD5 results not from 503F itself, but from one or more nearby hitchhiking variants, in the genes IRF1 or IL5. To test for evidence of recent positive selection on the 503F allele, we employed the iHS statistic, which was significant in the European CEU HapMap population (P=0.0007) and European Human Genome Diversity Panel populations (P≤0.01). To evaluate the hypothesis of disease-variant hitchhiking, we performed haplotype association tests on high-density microarray data in a sample of 1,868 Crohn's disease cases and 5,550 controls. We found that 503F haplotypes with recombination breakpoints between OCTN1 and IRF1 or IL5 were not associated with disease (odds ratio [OR]: 1.05, P=0.21). In contrast, we observed strong disease association for 503F haplotypes with no recombination between these three genes (OR: 1.24, P=2.6×10(-8)), as expected if the sweeping haplotype harbored one or more disease-causing mutations in IRF1 or IL5. To further evaluate these disease-gene candidates, we obtained expression data from lower gastrointestinal biopsies of healthy individuals and Crohn's disease patients. We observed a 72% increase in gene expression of IRF1 among Crohn's disease patients (P=0.0006) and no significant difference in expression of OCTN1. Collectively, these data indicate that the 503F variant has increased in frequency due to recent positive selection and that disease-causing variants in linkage disequilibrium with 503F have hitchhiked to relatively high frequency, thus forming the IBD5 risk haplotype. Finally, our association results and expression data support IRF1 as a strong candidate for Crohn's disease causation.
Figures



References
-
- Badr A, Muller K, Schafer-Pregl R, El Rabey H, Effgen S, Ibrahim HH, Pozzi C, Rohde W, Salamini F. On the origin and domestication history of Barley (Hordeum vulgare) Mol Biol Evol. 2000;17:499–510. - PubMed
-
- Bamshad MJ, Watkins WS, Dixon ME, Jorde LB, Rao BB, Naidu JM, Prasad BV, Rasanayagam A, Hammer MF. Female gene flow stratifies Hindu castes. Nature. 1998;395:651–652. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K23 DK069513/DK/NIDDK NIH HHS/United States
- DK069513/DK/NIDDK NIH HHS/United States
- 1P30DK078392-01/DK/NIDDK NIH HHS/United States
- R01 DK068164/DK/NIDDK NIH HHS/United States
- P30 DK078392/DK/NIDDK NIH HHS/United States
- C06 RR011234/RR/NCRR NIH HHS/United States
- CO6-RR11234/CO/NCI NIH HHS/United States
- T32 HL105321/HL/NHLBI NIH HHS/United States
- R01 DK078683/DK/NIDDK NIH HHS/United States
- GM59290/GM/NIGMS NIH HHS/United States
- UL1-RR025764/RR/NCRR NIH HHS/United States
- UL1 RR025764/RR/NCRR NIH HHS/United States
- R01 GM059290/GM/NIGMS NIH HHS/United States
- K99 HG005846/HG/NHGRI NIH HHS/United States
- T32 GM007464/GM/NIGMS NIH HHS/United States
- UL1 TR000077/TR/NCATS NIH HHS/United States
- K99HG005846/HG/NHGRI NIH HHS/United States
- 1T32HL105321-01/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

