Status and prospects for discovery and verification of new biomarkers of cardiovascular disease by proteomics
- PMID: 21817166
- PMCID: PMC3973157
- DOI: 10.1161/CIRCRESAHA.110.225003
Status and prospects for discovery and verification of new biomarkers of cardiovascular disease by proteomics
Abstract
Despite unmet needs for cardiovascular biomarkers, few new protein markers have been approved by the US Food and Drug Administration for the diagnosis or screening of cardiovascular diseases. Mass spectrometry-based proteomics technologies are capable of identifying hundreds to thousands of proteins in cells, tissues, and biofluids. Proteomics may therefore provide the opportunity to elucidate new biomarkers and pathways without a prior known association with cardiovascular disease; however, important obstacles remain. In this review, we focus on emerging techniques that may form a coherently integrated pipeline to overcome present limitations to both the discovery and validation processes.
Conflict of interest statement
Figures


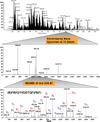
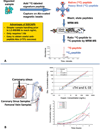
References
-
- Khot UN, Khot MB, Bajzer CT, Sapp SK, Ohman EM, Brener SJ, Ellis SG, Lincoff AM, Topol EJ. Prevalence of conventional risk factors in patients with coronary heart disease. JAMA. 2003;290:898–904. - PubMed
-
- Antman E, Grudzien C, Sacks DB. Evaluation of a rapid bedside assay for detection of serum cardiac troponin T. JAMA. 1995;273:1279–1282. - PubMed
-
- Lindahl B, Toss H, Siegbahn A, Venge P, Wallentin L for the FRISC Study Group. Fragmin during Instability in Coronary Artery Disease. Markers of myocardial damage and inflammation in relation to long-term mortality in unstable coronary artery disease. N Engl J Med. 2000;343:1139–1147. - PubMed
-
- Antman EM, Tanasijevic MJ, Thompson B, Schactman M, McCabe CH, Cannon CP, Fischer GA, Fung AY, Thompson C, Wybenga D, Braunwald E. Cardiac-specific troponin I levels to predict the risk of mortality in patients with acute coronary syndromes. N Engl J Med. 1996;335:1342–1349. - PubMed
-
- Hamm CW, Heeschen C, Goldmann B, Vahanian A, Adgey J, Miguel CM, Rutsch W, Berger J, Kootstra J, Simoons ML for the c7E3 Fab Antiplatelet Therapy in Unstable Refractory Angina (CAPTURE) Study Investigators. Benefit of abciximab in patients with refractory unstable angina in relation to serum troponin T levels. N. Engl. J. Med. 1999;340:1623–1629. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

