ENZO: a web tool for derivation and evaluation of kinetic models of enzyme catalyzed reactions
- PMID: 21818304
- PMCID: PMC3139599
- DOI: 10.1371/journal.pone.0022265
ENZO: a web tool for derivation and evaluation of kinetic models of enzyme catalyzed reactions
Abstract
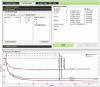
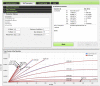
We describe a web tool ENZO (Enzyme Kinetics), a graphical interface for building kinetic models of enzyme catalyzed reactions. ENZO automatically generates the corresponding differential equations from a stipulated enzyme reaction scheme. These differential equations are processed by a numerical solver and a regression algorithm which fits the coefficients of differential equations to experimentally observed time course curves. ENZO allows rapid evaluation of rival reaction schemes and can be used for routine tests in enzyme kinetics. It is freely available as a web tool, at http://enzo.cmm.ki.si.
Conflict of interest statement
Figures






Similar articles
-
Automatic Assembly and Calibration of Models of Enzymatic Reactions Based on Ordinary Differential Equations.Methods Mol Biol. 2022;2385:141-152. doi: 10.1007/978-1-0716-1767-0_7. Methods Mol Biol. 2022. PMID: 34888719
-
Non-productive binding of butyryl(thio)choline in the active site of vertebrate acetylcholinesterase.Chem Biol Interact. 2010 Sep 6;187(1-3):128-34. doi: 10.1016/j.cbi.2010.05.001. Epub 2010 May 7. Chem Biol Interact. 2010. PMID: 20452336
-
interferENZY: A Web-Based Tool for Enzymatic Assay Validation and Standardized Kinetic Analysis.J Mol Biol. 2021 May 28;433(11):166613. doi: 10.1016/j.jmb.2020.07.025. Epub 2020 Aug 5. J Mol Biol. 2021. PMID: 32768452
-
Practical steady-state enzyme kinetics.Methods Enzymol. 2014;536:3-15. doi: 10.1016/B978-0-12-420070-8.00001-5. Methods Enzymol. 2014. PMID: 24423262 Review.
-
Fitting enzyme kinetic data with KinTek Global Kinetic Explorer.Methods Enzymol. 2009;467:601-626. doi: 10.1016/S0076-6879(09)67023-3. Methods Enzymol. 2009. PMID: 19897109 Review.
Cited by
-
Development of an in-vivo active reversible butyrylcholinesterase inhibitor.Sci Rep. 2016 Dec 21;6:39495. doi: 10.1038/srep39495. Sci Rep. 2016. PMID: 28000737 Free PMC article.
-
Investigation of Paraoxonase-1 Genotype and Enzyme-Kinetic Parameters in the Context of Cognitive Impairment in Parkinson's Disease.Antioxidants (Basel). 2023 Feb 7;12(2):399. doi: 10.3390/antiox12020399. Antioxidants (Basel). 2023. PMID: 36829958 Free PMC article.
-
The Removal of Time-Concentration Data Points from Progress Curves Improves the Determination of Km: The Example of Paraoxonase 1.Molecules. 2022 Feb 15;27(4):1306. doi: 10.3390/molecules27041306. Molecules. 2022. PMID: 35209091 Free PMC article.
-
Rapid modeling of experimental molecular kinetics with simple electronic circuits instead of with complex differential equations.Front Bioeng Biotechnol. 2022 Sep 28;10:947508. doi: 10.3389/fbioe.2022.947508. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36246369 Free PMC article.
-
Kinetically-controlled mechanism-based isolation of metabolic serine hydrolases in active form from complex proteomes: butyrylcholinesterase as a case study.RSC Adv. 2019 Nov 26;9(66):38505-38519. doi: 10.1039/c9ra07583f. eCollection 2019 Nov 25. RSC Adv. 2019. PMID: 35540231 Free PMC article.
References
-
- Michaelis L, Menten ML. Kinetik der Invertinwirkung. Biochem Z. 1913;49:333–369.
-
- Rosenberry TL. Acetylcholinesterase. Adv Enzymol. 1975;43:103–218. - PubMed
-
- Lineweaver H, Burk D. The determination of enzyme dissociation constants. J Am Chem Soc. 1934;56:658–666.
-
- Duggleby RG. Analysis of enzyme progress curves by non-linear regression. Methods Enzymol. 1995;249:61–90. - PubMed
-
- Kuzmic P. DynaFit–a software package for enzymology. Methods Enzymol. 2009;467:247–280. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

