A retrosynthetic biology approach to metabolic pathway design for therapeutic production
- PMID: 21819595
- PMCID: PMC3163555
- DOI: 10.1186/1752-0509-5-122
A retrosynthetic biology approach to metabolic pathway design for therapeutic production
Abstract
Background: Synthetic biology is used to develop cell factories for production of chemicals by constructively importing heterologous pathways into industrial microorganisms. In this work we present a retrosynthetic approach to the production of therapeutics with the goal of developing an in situ drug delivery device in host cells. Retrosynthesis, a concept originally proposed for synthetic chemistry, iteratively applies reversed chemical transformations (reversed enzyme-catalyzed reactions in the metabolic space) starting from a target product to reach precursors that are endogenous to the chassis. So far, a wider adoption of retrosynthesis into the manufacturing pipeline has been hindered by the complexity of enumerating all feasible biosynthetic pathways for a given compound.
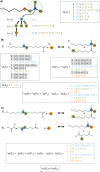
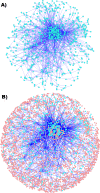
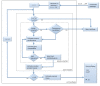
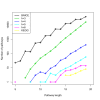
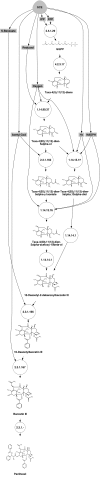
Results: In our method, we efficiently address the complexity problem by coding substrates, products and reactions into molecular signatures. Metabolic maps are represented using hypergraphs and the complexity is controlled by varying the specificity of the molecular signature. Furthermore, our method enables candidate pathways to be ranked to determine which ones are best to engineer. The proposed ranking function can integrate data from different sources such as host compatibility for inserted genes, the estimation of steady-state fluxes from the genome-wide reconstruction of the organism's metabolism, or the estimation of metabolite toxicity from experimental assays. We use several machine-learning tools in order to estimate enzyme activity and reaction efficiency at each step of the identified pathways. Examples of production in bacteria and yeast for two antibiotics and for one antitumor agent, as well as for several essential metabolites are outlined.
Conclusions: We present here a unified framework that integrates diverse techniques involved in the design of heterologous biosynthetic pathways through a retrosynthetic approach in the reaction signature space. Our engineering methodology enables the flexible design of industrial microorganisms for the efficient on-demand production of chemical compounds with therapeutic applications.
Figures
Similar articles
-
Retrosynthetic design of heterologous pathways.Methods Mol Biol. 2013;985:149-73. doi: 10.1007/978-1-62703-299-5_9. Methods Mol Biol. 2013. PMID: 23417804
-
Enumerating metabolic pathways for the production of heterologous target chemicals in chassis organisms.BMC Syst Biol. 2012 Feb 6;6:10. doi: 10.1186/1752-0509-6-10. BMC Syst Biol. 2012. PMID: 22309974 Free PMC article.
-
Construction and optimization of synthetic pathways in metabolic engineering.Curr Opin Microbiol. 2010 Jun;13(3):363-70. doi: 10.1016/j.mib.2010.02.004. Epub 2010 Mar 10. Curr Opin Microbiol. 2010. PMID: 20219419 Review.
-
Prototyping of microbial chassis for the biomanufacturing of high-value chemical targets.Biochem Soc Trans. 2021 Jun 30;49(3):1055-1063. doi: 10.1042/BST20200017. Biochem Soc Trans. 2021. PMID: 34100907 Review.
-
A retrosynthetic biology approach to therapeutics: from conception to delivery.Curr Opin Biotechnol. 2012 Dec;23(6):948-56. doi: 10.1016/j.copbio.2012.03.009. Epub 2012 Apr 2. Curr Opin Biotechnol. 2012. PMID: 22475981 Review.
Cited by
-
A Combinatorial Algorithm for Microbial Consortia Synthetic Design.Sci Rep. 2016 Jul 4;6:29182. doi: 10.1038/srep29182. Sci Rep. 2016. PMID: 27373593 Free PMC article.
-
A new network representation of the metabolism to detect chemical transformation modules.BMC Bioinformatics. 2015 Nov 14;16:385. doi: 10.1186/s12859-015-0809-4. BMC Bioinformatics. 2015. PMID: 26573681 Free PMC article.
-
Engineering biological systems using automated biofoundries.Metab Eng. 2017 Jul;42:98-108. doi: 10.1016/j.ymben.2017.06.003. Epub 2017 Jun 7. Metab Eng. 2017. PMID: 28602523 Free PMC article. Review.
-
dGPredictor: Automated fragmentation method for metabolic reaction free energy prediction and de novo pathway design.PLoS Comput Biol. 2021 Sep 27;17(9):e1009448. doi: 10.1371/journal.pcbi.1009448. eCollection 2021 Sep. PLoS Comput Biol. 2021. PMID: 34570771 Free PMC article.
-
Development of bio-based fine chemical production through synthetic bioengineering.Microb Cell Fact. 2014 Dec 14;13:173. doi: 10.1186/s12934-014-0173-5. Microb Cell Fact. 2014. PMID: 25494636 Free PMC article.
References
-
- Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MCY, Withers ST, Shiba Y, Sarpong R, Keasling JD. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440(7086):940–943. doi: 10.1038/nature04640. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

