Transcription analysis on response of swine lung to H1N1 swine influenza virus
- PMID: 21819625
- PMCID: PMC3169531
- DOI: 10.1186/1471-2164-12-398
Transcription analysis on response of swine lung to H1N1 swine influenza virus
Abstract
Background: As a mild, highly contagious, respiratory disease, swine influenza always damages the innate immune systems, and increases susceptibility to secondary infections which results in considerable morbidity and mortality in pigs. Nevertheless, the systematical host response of pigs to swine influenza virus infection remains largely unknown. To explore it, a time-course gene expression profiling was performed for comprehensive analysis of the global host response induced by H1N1 swine influenza virus in pigs.
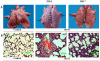
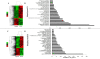
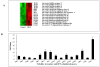
Results: At the early stage of H1N1 swine virus infection, pigs were suffering mild respiratory symptoms and pathological changes. A total of 268 porcine genes showing differential expression (DE) after inoculation were identified to compare with the controls on day 3 post infection (PID) (Fold change ≥ 2, p < 0.05). The DE genes were involved in many vital functional classes, mainly including signal transduction, immune response, inflammatory response, cell adhesion and cell-cell signalling. Noticeably, the genes associated with immune and inflammatory response showed highly overexpressed. Through the pathway analysis, the significant pathways mainly concerned with Cell adhesion molecules, Cytokine-cytokine receptor interaction, Toll-like receptor signaling pathway and MAPK signaling pathway, suggesting that the host took different strategies to activate these pathways so as to prevent virus infections at the early stage. However, on PID 7, the predominant function classes of DE genes included signal transduction, metabolism, transcription, development and transport. Furthermore, the most significant pathways switched to PPAR signaling pathway and complement and coagulation cascades, showing that the host might start to repair excessive tissue damage by anti-inflammatory functions. These results on PID 7 demonstrated beneficial turnover for host to prevent excessive inflammatory damage and recover the normal state by activating these clusters of genes.
Conclusions: This study shows how the target organ responds to H1N1 swine influenza virus infection in pigs. The observed gene expression profile could help to screen the potential host agents for reducing the prevalence of swine influenza virus and further understand the molecular pathogenesis associated with H1N1 infection in pigs.
Figures
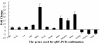
Similar articles
-
Investigation of Pathogenesis of H1N1 Influenza Virus and Swine Streptococcus suis Serotype 2 Co-Infection in Pigs by Microarray Analysis.PLoS One. 2015 Apr 23;10(4):e0124086. doi: 10.1371/journal.pone.0124086. eCollection 2015. PLoS One. 2015. PMID: 25906258 Free PMC article.
-
2009 pandemic H1N1 influenza virus elicits similar clinical course but differential host transcriptional response in mouse, macaque, and swine infection models.BMC Genomics. 2012 Nov 15;13:627. doi: 10.1186/1471-2164-13-627. BMC Genomics. 2012. PMID: 23153050 Free PMC article.
-
2009 pandemic H1N1 influenza virus causes disease and upregulation of genes related to inflammatory and immune responses, cell death, and lipid metabolism in pigs.J Virol. 2011 Nov;85(22):11626-37. doi: 10.1128/JVI.05705-11. Epub 2011 Sep 7. J Virol. 2011. PMID: 21900171 Free PMC article.
-
[Swine influenza virus: evolution mechanism and epidemic characterization--a review].Wei Sheng Wu Xue Bao. 2009 Sep;49(9):1138-45. Wei Sheng Wu Xue Bao. 2009. PMID: 20030049 Review. Chinese.
-
Isolation and genetic characterization of avian-like H1N1 and novel ressortant H1N2 influenza viruses from pigs in China.Biochem Biophys Res Commun. 2009 Aug 21;386(2):278-83. doi: 10.1016/j.bbrc.2009.05.056. Epub 2009 May 19. Biochem Biophys Res Commun. 2009. PMID: 19460353 Review.
Cited by
-
Nonstructural protein 1 of influenza A virus interacts with human guanylate-binding protein 1 to antagonize antiviral activity.PLoS One. 2013;8(2):e55920. doi: 10.1371/journal.pone.0055920. Epub 2013 Feb 6. PLoS One. 2013. PMID: 23405236 Free PMC article.
-
Conjoint analysis of transcriptome and metabolome profiles of normal captivity and arch soil free-range in Meishan pigs.Front Vet Sci. 2023 Jul 27;10:1187877. doi: 10.3389/fvets.2023.1187877. eCollection 2023. Front Vet Sci. 2023. PMID: 37576833 Free PMC article.
-
Transcriptomic and Epigenetic Profiling of the Lung of Influenza-Infected Pigs: A Comparison of Different Birth Weight and Susceptibility Groups.PLoS One. 2015 Sep 22;10(9):e0138653. doi: 10.1371/journal.pone.0138653. eCollection 2015. PLoS One. 2015. PMID: 26393920 Free PMC article.
-
Transcription analysis on response of porcine alveolar macrophages to Haemophilus parasuis.BMC Genomics. 2012 Feb 13;13:68. doi: 10.1186/1471-2164-13-68. BMC Genomics. 2012. PMID: 22330747 Free PMC article.
-
Development of an objective gene expression panel as an alternative to self-reported symptom scores in human influenza challenge trials.J Transl Med. 2017 Jun 8;15(1):134. doi: 10.1186/s12967-017-1235-3. J Transl Med. 2017. PMID: 28595644 Free PMC article. Clinical Trial.
References
-
- Van Reeth K, Nauwynck H, Pensaert M. Clinical effects of experimental dual infections with porcine reproductive and respiratory syndrome virus followed by swine influenza virus in conventional and colostrum-deprived pigs. J Vet Med. 2001;48(4):283–292. doi: 10.1046/j.1439-0450.2001.00438.x. - DOI - PMC - PubMed
-
- Brown IH, Ludwig S, Olsen CW, Hannoun C, Scholtissek C, Hinshaw VS, Harris PA, McCauley JW, Strong I, Alexander DJ. Antigenic and genetic analyses of H1N1 influenza A viruses from European pigs. J Gen Virol. 1997;78(3):553–562. - PubMed
-
- Richt JA, Lager KM, Janke BH, Woods RD, Webster RG, Webby RJ. Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States. J Clin Microbial. 2003;41(7):3198–3205. doi: 10.1128/JCM.41.7.3198-3205.2003. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

