Highly sensitive detection of S-nitrosylated proteins by capillary gel electrophoresis with laser induced fluorescence
- PMID: 21820121
- PMCID: PMC3164759
- DOI: 10.1016/j.chroma.2011.07.062
Highly sensitive detection of S-nitrosylated proteins by capillary gel electrophoresis with laser induced fluorescence
Abstract
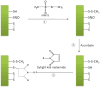
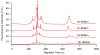
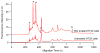
S-nitrosylated proteins are biomarkers of oxidative damage in aging and Alzheimer's disease (AD). Here, we report a new method for detecting and quantifying nitrosylated proteins by capillary gel electrophoresis with laser induced fluorescence detection (CGE-LIF). Dylight 488 maleimide was used to specifically label thiol group (SH) after switching the S-nitrosothiol (S-NO) to SH in cysteine using the "fluorescence switch" assay. In vitro nitrosylation model-BSA subjected to S-nitrosoglutathione (GSNO) optimized the labeling reactions and characterized the response of the LIF detector. The method proves to be highly sensitive, detecting 1.3 picomolar (pM) concentration of nitrosothiols in nanograms of proteins, which is the lowest limit of detection of nitrosothiols reported to date. We further demonstrated the direct application of this method in monitoring protein nitrosylation damage in MQ mediated human colon adenocarcinoma cells. The nitrosothiol amounts in MQ treated and untreated cells are 14.8±0.2 and 10.4±0.5 pmol/mg of proteins, respectively. We also depicted nitrosylated protein electrophoretic profiles of brain cerebrum of 5-month-old AD transgenic (Tg) mice model. In Tg mice brain, 15.5±0.4 pmol of nitrosothiols/mg of proteins was quantified while wild type contained 11.7±0.3 pmol/mg proteins. The methodology is validated to quantify low levels of S-nitrosylated protein in complex protein mixtures from both physiological and pathological conditions.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures


Similar articles
-
S-Glutathionyl quantification in the attomole range using glutaredoxin-3-catalyzed cysteine derivatization and capillary gel electrophoresis with laser-induced fluorescence detection.Anal Bioanal Chem. 2011 Oct;401(7):2165-75. doi: 10.1007/s00216-011-5311-x. Epub 2011 Aug 13. Anal Bioanal Chem. 2011. PMID: 21842197 Free PMC article.
-
Two-dimensional nitrosylated protein fingerprinting by using poly (methyl methacrylate) microchips.Lab Chip. 2012 Sep 21;12(18):3362-9. doi: 10.1039/c2lc40132k. Epub 2012 Jul 6. Lab Chip. 2012. PMID: 22766561
-
A "fluorescence switch" technique increases the sensitivity of proteomic detection and identification of S-nitrosylated proteins.Proteomics. 2009 Dec;9(23):5359-70. doi: 10.1002/pmic.200900070. Proteomics. 2009. PMID: 19798666
-
Measurement of protein S-nitrosylation during cell signaling.Methods Enzymol. 2008;440:231-42. doi: 10.1016/S0076-6879(07)00814-2. Methods Enzymol. 2008. PMID: 18423221 Review.
-
S-Nitrosylation in Alzheimer's disease.Mol Neurobiol. 2015 Feb;51(1):268-80. doi: 10.1007/s12035-014-8672-2. Epub 2014 Mar 25. Mol Neurobiol. 2015. PMID: 24664522 Review.
Cited by
-
The roles of S-nitrosylation and S-glutathionylation in Alzheimer's disease.Methods Enzymol. 2019;626:499-538. doi: 10.1016/bs.mie.2019.08.004. Methods Enzymol. 2019. PMID: 31606089 Free PMC article. Review.
-
Proteomic approaches to quantify cysteine reversible modifications in aging and neurodegenerative diseases.Proteomics Clin Appl. 2016 Dec;10(12):1159-1177. doi: 10.1002/prca.201600015. Epub 2016 Nov 11. Proteomics Clin Appl. 2016. PMID: 27666938 Free PMC article. Review.
-
Yin and Yang in Post-Translational Modifications of Human D-Amino Acid Oxidase.Front Mol Biosci. 2021 May 10;8:684934. doi: 10.3389/fmolb.2021.684934. eCollection 2021. Front Mol Biosci. 2021. PMID: 34041270 Free PMC article.
References
-
- Martinez-Ruiz A, Lamas S. Cardiovasc Res. 2004;62:43. - PubMed
-
- Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Nat Cell Biol. 2001;3:193. - PubMed
-
- Huber SC, Hardin SC. Curr Opin Plant Biol. 2004;7:318. - PubMed
-
- Spickett CM, Pitt AR, Morrice N, Kolch W. Biochim Biophys Acta. 2006;1764:1823. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

