Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome
- PMID: 21820332
- PMCID: PMC3162998
- DOI: 10.1016/j.immuni.2011.05.016
Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome
Abstract
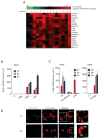
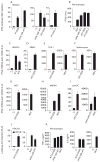
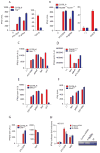
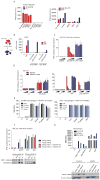
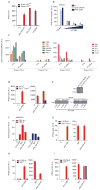
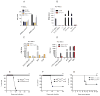
Although Toll-like receptor 9 (TLR9) has been implicated in cytokine and type I interferon (IFN) production during malaria in humans and mice, the high AT content of the Plasmodium falciparum genome prompted us to examine the possibility that malarial DNA triggered TLR9-independent pathways. Over 6000 ATTTTTAC ("AT-rich") motifs are present in the genome of P. falciparum, which we show here potently induce type I IFNs. Parasite DNA, parasitized erythrocytes and oligonucleotides containing the AT-rich motif induce type I IFNs via a pathway that did not involve the previously described sensors TLR9, DAI, RNA polymerase-III or IFI16/p204. Rather, AT-rich DNA sensing involved an unknown receptor that coupled to the STING, TBK1 and IRF3-IRF7 signaling pathway. Mice lacking IRF3, IRF7, the kinase TBK1 or the type I IFN receptor were resistant to otherwise lethal cerebral malaria. Collectively, these observations implicate AT-rich DNA sensing via STING, TBK1 and IRF3-IRF7 in P. falciparum malaria.
Copyright © 2011 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Comment in
-
Innate immunity: AT-rich DNA trapped in the cytoplasm.Nat Rev Immunol. 2011 Aug 25;11(9):569. doi: 10.1038/nri3057. Nat Rev Immunol. 2011. PMID: 21866165 No abstract available.
-
Taking the sting out of malaria.Immunity. 2011 Aug 26;35(2):149-51. doi: 10.1016/j.immuni.2011.08.002. Immunity. 2011. PMID: 21867921
References
-
- Adachi K, Tsutsui H, Kashiwamura S, Seki E, Nakano H, Takeuchi O, Takeda K, Okumura K, Van Kaer L, Okamura H, et al. Plasmodium berghei infection in mice induces liver injury by an IL-12- and toll-like receptor/myeloid differentiation factor 88-dependent mechanism. J Immunol. 2001;167:5928–5934. - PubMed
-
- Aucan C, Walley AJ, Hennig BJ, Fitness J, Frodsham A, Zhang L, Kwiatkowski D, Hill AV. Interferon-alpha receptor-1 (IFNAR1) variants are associated with protection against cerebral malaria in the Gambia. Genes Immun. 2003;4:275–282. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

