The Staphylococcus aureus pathogenicity island 1 protein gp6 functions as an internal scaffold during capsid size determination
- PMID: 21821042
- PMCID: PMC3175317
- DOI: 10.1016/j.jmb.2011.07.036
The Staphylococcus aureus pathogenicity island 1 protein gp6 functions as an internal scaffold during capsid size determination
Abstract
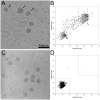
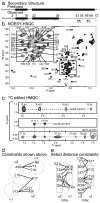
Staphylococcus aureus pathogenicity island 1 (SaPI1) is a mobile genetic element that carries genes for several superantigen toxins. SaPI1 is normally stably integrated into the host genome but can become mobilized by "helper" bacteriophage 80α, leading to the packaging of SaPI1 genomes into phage-like transducing particles that are composed of structural proteins supplied by the helper phage but having smaller capsids. We show that the SaPI1-encoded protein gp6 is necessary for efficient formation of small capsids. The NMR structure of gp6 reveals a dimeric protein with a helix-loop-helix motif similar to that of bacteriophage scaffolding proteins. The gp6 dimer matches internal densities that bridge capsid subunits in cryo-electron microscopy reconstructions of SaPI1 procapsids, suggesting that gp6 acts as an internal scaffolding protein in capsid size determination.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures



References
-
- Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3:722–732. - PubMed
-
- Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

