Structural constraints in collaborative competition of transcription factors against the nucleosome
- PMID: 21821044
- PMCID: PMC3534743
- DOI: 10.1016/j.jmb.2011.07.032
Structural constraints in collaborative competition of transcription factors against the nucleosome
Abstract
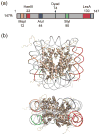
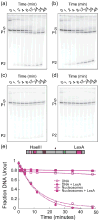
Cooperativity in transcription factor (TF) binding is essential in eukaryotic gene regulation and arises through diverse mechanisms. Here, we focus on one mechanism, collaborative competition, which is of interest because it arises both automatically (with no requirement for TF coevolution) and spontaneously (with no requirement for ATP-dependent nucleosome remodeling factors). Previous experimental studies of collaborative competition analyzed cases in which target sites for pairs of cooperating TFs were contained within the same side of the nucleosome. Here, we utilize new assays to measure cooperativity in protein binding to pairs of nucleosomal DNA target sites. We focus on the cases that are of greatest in vivo relevance, in which one binding site is located close to the end of a nucleosome and the other binding site is located at diverse positions throughout the nucleosome. Our results reveal energetically significant positive (favorable) cooperativity for pairs of sites on the same side of the nucleosome but, for the cases examined, energetically insignificant cooperativity between sites on opposite sides of the nucleosome. These findings imply a special significance for TF binding sites that are spaced within one-half nucleosome length (74 bp) or less along the genome and may prove useful for prediction of cooperatively acting TFs genome wide.
Copyright © 2011. Published by Elsevier Ltd.
Figures



Similar articles
-
Identifying cooperative transcription factors in yeast using multiple data sources.BMC Syst Biol. 2014;8 Suppl 5(Suppl 5):S2. doi: 10.1186/1752-0509-8-S5-S2. Epub 2014 Dec 12. BMC Syst Biol. 2014. PMID: 25559499 Free PMC article.
-
Nucleosome-mediated cooperativity between transcription factors.Proc Natl Acad Sci U S A. 2010 Dec 28;107(52):22534-9. doi: 10.1073/pnas.0913805107. Epub 2010 Dec 13. Proc Natl Acad Sci U S A. 2010. PMID: 21149679 Free PMC article.
-
Nucleosomal context of binding sites influences transcription factor binding affinity and gene regulation.Genomics Proteomics Bioinformatics. 2009 Dec;7(4):155-62. doi: 10.1016/S1672-0229(08)60045-5. Genomics Proteomics Bioinformatics. 2009. PMID: 20172488 Free PMC article.
-
Epigenome Regulation by Dynamic Nucleosome Unwrapping.Trends Biochem Sci. 2020 Jan;45(1):13-26. doi: 10.1016/j.tibs.2019.09.003. Epub 2019 Oct 17. Trends Biochem Sci. 2020. PMID: 31630896 Free PMC article. Review.
-
Changing the DNA landscape: putting a SPN on chromatin.Curr Top Microbiol Immunol. 2003;274:171-201. doi: 10.1007/978-3-642-55747-7_7. Curr Top Microbiol Immunol. 2003. PMID: 12596908 Review.
Cited by
-
Codons support the maintenance of intrinsic DNA polymer flexibility over evolutionary timescales.Genome Biol Evol. 2012;4(9):954-65. doi: 10.1093/gbe/evs073. Epub 2012 Aug 30. Genome Biol Evol. 2012. PMID: 22936074 Free PMC article.
-
Ensembles of Breathing Nucleosomes: A Computational Study.Biophys J. 2020 May 5;118(9):2297-2308. doi: 10.1016/j.bpj.2019.11.3395. Epub 2019 Dec 12. Biophys J. 2020. PMID: 31882248 Free PMC article.
-
Functional roles of nucleosome stability and dynamics.Brief Funct Genomics. 2015 Jan;14(1):50-60. doi: 10.1093/bfgp/elu038. Epub 2014 Sep 30. Brief Funct Genomics. 2015. PMID: 25275099 Free PMC article. Review.
-
Natural Genetic Variation Reveals Key Features of Epigenetic and Transcriptional Memory in Virus-Specific CD8 T Cells.Immunity. 2019 May 21;50(5):1202-1217.e7. doi: 10.1016/j.immuni.2019.03.031. Epub 2019 Apr 23. Immunity. 2019. PMID: 31027997 Free PMC article.
-
Quantitative Modeling of Nucleosome Unwrapping from Both Ends.Biophys J. 2019 Dec 3;117(11):2204-2216. doi: 10.1016/j.bpj.2019.09.048. Epub 2019 Oct 30. Biophys J. 2019. PMID: 31732143 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

