plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics
- PMID: 21821130
- PMCID: PMC3298692
- DOI: 10.1016/j.jsb.2011.07.009
plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics
Abstract
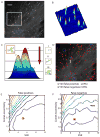
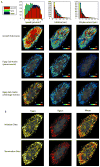
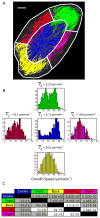
Here we introduce plusTipTracker, a Matlab-based open source software package that combines automated tracking, data analysis, and visualization tools for movies of fluorescently-labeled microtubule (MT) plus end binding proteins (+TIPs). Although +TIPs mark only phases of MT growth, the plusTipTracker software allows inference of additional MT dynamics, including phases of pause and shrinkage, by linking collinear, sequential growth tracks. The algorithm underlying the reconstruction of full MT trajectories relies on the spatially and temporally global tracking framework described in Jaqaman et al. (2008). Post-processing of track populations yields a wealth of quantitative phenotypic information about MT network architecture that can be explored using several visualization modalities and bioinformatics tools included in plusTipTracker. Graphical user interfaces enable novice Matlab users to track thousands of MTs in minutes. In this paper, we describe the algorithms used by plusTipTracker and show how the package can be used to study regional differences in the relative proportion of MT subpopulations within a single cell. The strategy of grouping +TIP growth tracks for the analysis of MT dynamics has been introduced before (Matov et al., 2010). The numerical methods and analytical functionality incorporated in plusTipTracker substantially advance this previous work in terms of flexibility and robustness. To illustrate the enhanced performance of the new software we thus compare computer-assembled +TIP-marked trajectories to manually-traced MT trajectories from the same movie used in Matov et al. (2010).
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Acquisition frame rate affects microtubule plus-end tracking analysis.Nat Methods. 2014 Mar;11(3):219-20. doi: 10.1038/nmeth.2846. Nat Methods. 2014. PMID: 24577268 No abstract available.
References
-
- Akhmanova A, Hoogenraad CC. Microtubule plus-end-tracking proteins: mechanisms and functions. Current Opinion in Cell Biology. 2005;17:47–54. - PubMed
-
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, et al. Molecular biology of the cell. 4. Garland Science; New York: 2002.
-
- Applegate KT, Danuser G. Technical Report: Using the plusTipTracker Software to Measure and Analyze Microtubule Dynamics from +TIP Comets. 2010 Accompanying software release ( lccb.hms.harvard.edu/software)
-
- Bieling P, Laan L, Schek H, Munteanu EL, Sandblad L, et al. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450:1100–1105. - PubMed
-
- Burkard KE, Cela E. Linear Assignment Problems and Extensions. In: Du DZ, Pardalos PM, editors. Handbook of Combinatorial Optimization - Supplement Volume A. Supp A. Kluwer Academic Publishers; Dordrecht, NL: 1999. pp. 75–149.pp. 75–149.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

