Salicylate functions as an efflux pump inducer and promotes the emergence of fluoroquinolone-resistant Campylobacter jejuni mutants
- PMID: 21821741
- PMCID: PMC3194847
- DOI: 10.1128/AEM.00763-11
Salicylate functions as an efflux pump inducer and promotes the emergence of fluoroquinolone-resistant Campylobacter jejuni mutants
Abstract
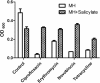
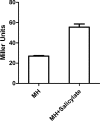
Salicylate, a nonsteroidal anti-inflammatory compound, has been shown to increase the resistance of Campylobacter to antimicrobials. However, the molecular mechanism underlying salicylate-induced resistance has not yet been established. In this study, we determined how salicylate increases antibiotic resistance and evaluated its impact on the development of fluoroquinolone-resistant Campylobacter mutants. Transcriptional fusion assays, real-time quantitative reverse transcription-PCR (RT-PCR), and immunoblotting assays consistently demonstrated the induction of the CmeABC multidrug efflux pump by salicylate. Electrophoretic mobility shift assays further showed that salicylate inhibits the binding of CmeR (a transcriptional repressor of the TetR family) to the promoter DNA of cmeABC, suggesting that salicylate inhibits the function of CmeR. The presence of salicylate in the culture medium not only decreased the susceptibility of Campylobacter to ciprofloxacin but also resulted in an approximately 70-fold increase in the observed frequency of emergence of fluoroquinolone-resistant mutants under selection with ciprofloxacin. Together, these results indicate that in Campylobacter, salicylate inhibits the binding of CmeR to the promoter DNA and induces expression of cmeABC, resulting in decreased susceptibility to antibiotics and in increased emergence of fluoroquinolone-resistant mutants under selection pressure.
Figures
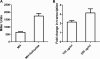


References
-
- Akiba M., Lin J., Barton Y. W., Zhang Q. 2006. Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J. Antimicrob. Chemother. 57:52–60 - PubMed
-
- Domenico P., Hopkins T., Cunha B. A. 1990. The effect of sodium salicylate on antibiotic susceptibility and synergy in Klebsiella pneumoniae. J. Antimicrob. Chemother. 26:343–351 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

