Matrix-assisted laser desorption ionization (MALDI)-time of flight mass spectrometry- and MALDI biotyper-based identification of cultured biphenyl-metabolizing bacteria from contaminated horseradish rhizosphere soil
- PMID: 21821747
- PMCID: PMC3187117
- DOI: 10.1128/AEM.05465-11
Matrix-assisted laser desorption ionization (MALDI)-time of flight mass spectrometry- and MALDI biotyper-based identification of cultured biphenyl-metabolizing bacteria from contaminated horseradish rhizosphere soil
Abstract
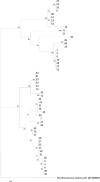
Bacteria that are able to utilize biphenyl as a sole source of carbon were extracted and isolated from polychlorinated biphenyl (PCB)-contaminated soil vegetated by horseradish. Isolates were identified using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). The usage of MALDI Biotyper for the classification of isolates was evaluated and compared to 16S rRNA gene sequence analysis. A wide spectrum of bacteria was isolated, with Arthrobacter, Serratia, Rhodococcus, and Rhizobium being predominant. Arthrobacter isolates also represented the most diverse group. The use of MALDI Biotyper in many cases permitted the identification at the level of species, which was not achieved by 16S rRNA gene sequence analyses. However, some isolates had to be identified by 16S rRNA gene analyses if MALDI Biotyper-based identification was at the level of probable or not reliable identification, usually due to a lack of reference spectra included in the database. Overall, this study shows the possibility of using MALDI-TOF MS and MALDI Biotyper for the fast and relatively nonlaborious identification/classification of soil isolates. At the same time, it demonstrates the dominant role of employing 16S rRNA gene analyses for the identification of recently isolated strains that can later fill the gaps in the protein-based identification databases.
Figures


References
-
- Abraham W. R., Wenderoth D. F., Glasser W. 2005. Diversity of biphenyl degraders in a chlorobenzene polluted aquifer. Chemosphere 58:529–533 - PubMed
-
- Alispahic M., et al. 2010. Species-specific identification and differentiation of Arcobacter, Helicobacter and Campylobacter by full-spectral matrix-associated laser desorption/ionization time of flight mass spectrometry analysis. J. Med. Microbiol. 59:295–301 - PubMed
-
- Arnold R. J., Reilly J. P. 1998. Fingerprint matching of E. coli strains with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry of whole cells using a modified correlation approach. Rapid Commun. Mass Spectrom. 12:630–636 - PubMed
-
- Asturias J. A., Diaz E., Timmis K. N. 1995. The evolutionary relationship of biphenyl dioxygenase from gram-positive Rhodococcus globerulus P6 to multicomponent dioxygenases from gram-negative bacteria. Gene 156:11–18 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

