Acquired antibiotic resistance: are we born with it?
- PMID: 21821748
- PMCID: PMC3194877
- DOI: 10.1128/AEM.05087-11
Acquired antibiotic resistance: are we born with it?
Abstract
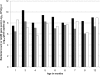
The rapid emergence of antibiotic resistance (AR) is a major public health concern. Recent findings on the prevalence of food-borne antibiotic-resistant (ART) commensal bacteria in ready-to-consume food products suggested that daily food consumption likely serves as a major avenue for dissemination of ART bacteria from the food chain to human hosts. To properly assess the impact of various factors, including the food chain, on AR development in hosts, it is important to determine the baseline of ART bacteria in the human gastrointestinal (GI) tract. We thus examined the gut microbiota of 16 infant subjects, from the newborn stage to 1 year of age, who fed on breast milk and/or infant formula during the early stages of development and had no prior exposure to antibiotics. Predominant bacterial populations resistant to several antibiotics and multiple resistance genes were found in the infant GI tracts within the first week of age. Several ART population transitions were also observed in the absence of antibiotic exposure and dietary changes. Representative AR gene pools including tet(M), ermB, sul2, and bla(TEM) were detected in infant subjects. Enterococcus spp., Staphylococcus spp., Klebsiella spp., Streptococcus spp., and Escherichia coli/Shigella spp. were among the identified AR gene carriers. ART bacteria were not detected in the infant formula and infant foods examined, but small numbers of skin-associated ART bacteria were found in certain breast milk samples. The data suggest that the early development of AR in the human gut microbiota is independent of infants' exposure to antibiotics but is likely impacted by exposure to maternal and environmental microbes during and after delivery and that the ART population is significantly amplified within the host even in the absence of antibiotic selective pressure.
Figures
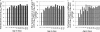


References
-
- Allen H. K., et al. 2010. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8:251–259 - PubMed
-
- Andremont A. 2003. Commensal flora may play key role in spreading antibiotic resistance. ASM News 69:601–607
-
- Bartoloni A., et al. 2004. High prevalence of acquired antimicrobial resistance unrelated to heavy antimicrobial consumption. J. Infect. Dis. 189:1291–1294 - PubMed
-
- Cabello F. C. 2006. Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ. Microbiol. 8:1137–1144 - PubMed
-
- Carmeli Y. 2008. Strategies for managing today's infections. Clin. Microbiol. Infect. 14(Suppl. 3):22–31 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

