Medicago truncatula CYP716A12 is a multifunctional oxidase involved in the biosynthesis of hemolytic saponins
- PMID: 21821776
- PMCID: PMC3180811
- DOI: 10.1105/tpc.111.087312
Medicago truncatula CYP716A12 is a multifunctional oxidase involved in the biosynthesis of hemolytic saponins
Abstract
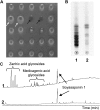
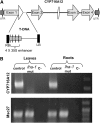
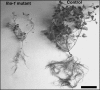
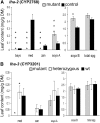
Saponins, a group of glycosidic compounds present in several plant species, have aglycone moieties that are formed using triterpenoid or steroidal skeletons. In spite of their importance as antimicrobial compounds and their possible benefits for human health, knowledge of the genetic control of saponin biosynthesis is still poorly understood. In the Medicago genus, the hemolytic activity of saponins is related to the nature of their aglycone moieties. We have identified a cytochrome P450 gene (CYP716A12) involved in saponin synthesis in Medicago truncatula using a combined genetic and biochemical approach. Genetic loss-of-function analysis and complementation studies showed that CYP716A12 is responsible for an early step in the saponin biosynthetic pathway. Mutants in CYP716A12 were unable to produce hemolytic saponins and only synthetized soyasaponins, and were thus named lacking hemolytic activity (lha). In vitro enzymatic activity assays indicate that CYP716A12 catalyzes the oxidation of β-amyrin and erythrodiol at the C-28 position, yielding oleanolic acid. Transcriptome changes in the lha mutant showed a modulation in the main steps of triterpenic saponin biosynthetic pathway: squalene cyclization, β-amyrin oxidation, and glycosylation. The analysis of CYP716A12 expression in planta is reported together with the sapogenin content in different tissues and stages. This article provides evidence for CYP716A12 being a key gene in hemolytic saponin biosynthesis.
Figures





References
-
- Achnine L., Huhman D.V., Farag M.A., Sumner L.W., Blount J.W., Dixon R.A. (2005). Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J. 41: 875–887 - PubMed
-
- Bialy Z., Jurzysta M., Mella M., Tava A. (2006). Triterpene saponins from the roots of Medicago hybrida. J. Agric. Food Chem. 54: 2520–2526 - PubMed
-
- Gamas P., de Billy F., Truchet G. (1998). Symbiosis-specific expression of two Medicago truncatula nodulin genes, MtN1 and MtN13, encoding products homologous to plant defense proteins. Mol. Plant Microbe Interact. 11: 393–403 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
- BB/E01772X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/B/1356X/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I001271/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E022758/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G17764/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

