Phosphatidylinositol monophosphate-binding interface in the oomycete RXLR effector AVR3a is required for its stability in host cells to modulate plant immunity
- PMID: 21821794
- PMCID: PMC3167543
- DOI: 10.1073/pnas.1106002108
Phosphatidylinositol monophosphate-binding interface in the oomycete RXLR effector AVR3a is required for its stability in host cells to modulate plant immunity
Abstract
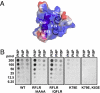
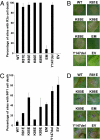
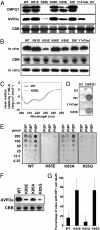
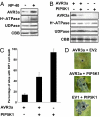
The oomycete pathogen Phytophthora infestans causes potato late blight, one of the most economically damaging plant diseases worldwide. P. infestans produces AVR3a, an essential modular virulence effector with an N-terminal RXLR domain that is required for host-cell entry. In host cells, AVR3a stabilizes and inhibits the function of the E3 ubiquitin ligase CMPG1, a key factor in host immune responses including cell death triggered by the pathogen-derived elicitor protein INF1 elicitin. To elucidate the molecular basis of AVR3a effector function, we determined the structure of Phytophthora capsici AVR3a4, a close homolog of P. infestans AVR3a. Our structural and functional analyses reveal that the effector domain of AVR3a contains a conserved, positively charged patch and that this region, rather than the RXLR domain, is required for binding to phosphatidylinositol monophosphates (PIPs) in vitro. Mutations affecting PIP binding do not abolish AVR3a recognition by the resistance protein R3a but reduce its ability to suppress INF1-triggered cell death in planta. Similarly, stabilization of CMPG1 in planta is diminished by these mutations. The steady-state levels of non-PIP-binding mutant proteins in planta are reduced greatly, although these proteins are stable in vitro. Furthermore, overexpression of a phosphatidylinositol phosphate 5-kinase results in reduction of AVR3a levels in planta. Our results suggest that the PIP-binding ability of the AVR3a effector domain is essential for its accumulation inside host cells to suppress CMPG1-dependent immunity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Showdown at the RXLR motif: Serious differences of opinion in how effector proteins from filamentous eukaryotic pathogens enter plant cells.Proc Natl Acad Sci U S A. 2011 Aug 30;108(35):14381-2. doi: 10.1073/pnas.1111668108. Epub 2011 Aug 19. Proc Natl Acad Sci U S A. 2011. PMID: 21856948 Free PMC article. No abstract available.
References
-
- Haverkort AJ, et al. Societal costs of late blight in potato and prospects of durable resistance through cisgenic modification. Potato Res. 2008;51:47–57.
-
- Haas BJ, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. - PubMed
-
- Kamoun S. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu Rev Phytopathol. 2006;44:41–60. - PubMed
-
- Oliva R, et al. Recent developments in effector biology of filamentous plant pathogens. Cell Microbiol. 2010;12:705–715. - PubMed
-
- Birch PRJ, et al. Oomycete RXLR effectors: Delivery, functional redundancy and durable disease resistance. Curr Opin Plant Biol. 2008;11:373–379. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

