Structure of filamin A immunoglobulin-like repeat 10 from Homo sapiens
- PMID: 21821884
- PMCID: PMC3151117
- DOI: 10.1107/S1744309111024249
Structure of filamin A immunoglobulin-like repeat 10 from Homo sapiens
Abstract
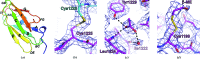
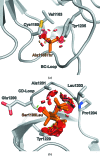
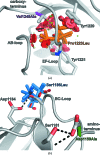
Filamin A (FlnA) plays a critical role in cytoskeletal organization, cell motility and cellular signaling. FlnA utilizes different binding sites on a series of 24 immunoglobulin-like domains (Ig repeats) to interact with diverse cytosolic proteins and with cytoplasmic portions of membrane proteins. Mutations in a specific domain, Ig10 (FlnA-Ig10), are correlated with two severe forms of the otopalatodigital syndrome spectrum disorders Melnick-Needles syndrome and frontometaphyseal dysplasia. The crystal structure of FlnA-Ig10 determined at 2.44 Å resolution provides insight into the perturbations caused by these mutations.
Figures

References
-
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
-
- Bork, P., Holm, L. & Sander, C. (1994). J. Mol. Biol. 242, 309–320. - PubMed
-
- DeLano, W. L. (2002). PyMOL http://www.pymol.org.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

