The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation
- PMID: 21822279
- PMCID: PMC4531310
- DOI: 10.1038/ncb2304
The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation
Abstract
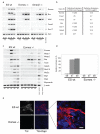
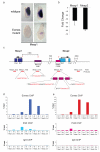
Instructive programmes guiding cell-fate decisions in the developing mouse embryo are controlled by a few so-termed master regulators. Genetic studies demonstrate that the T-box transcription factor Eomesodermin (Eomes) is essential for epithelial-to-mesenchymal transition, mesoderm migration and specification of definitive endoderm during gastrulation. Here we report that Eomes expression within the primitive streak marks the earliest cardiac mesoderm and promotes formation of cardiovascular progenitors by directly activating the bHLH (basic-helix-loop-helix) transcription factor gene Mesp1 upstream of the core cardiac transcriptional machinery. In marked contrast to Eomes/Nodal signalling interactions that cooperatively regulate anterior-posterior axis patterning and allocation of the definitive endoderm cell lineage, formation of cardiac progenitors requires only low levels of Nodal activity accomplished through a Foxh1/Smad4-independent mechanism. Collectively, our experiments demonstrate that Eomes governs discrete context-dependent transcriptional programmes that sequentially specify cardiac and definitive endoderm progenitors during gastrulation.
Figures



References
-
- Bondue A, et al. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. - PubMed
-
- David R, et al. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat Cell Biol. 2008;10:338–345. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

