Use of nucleic Acid analogs for the study of nucleic Acid interactions
- PMID: 21822475
- PMCID: PMC3142669
- DOI: 10.4061/2011/967098
Use of nucleic Acid analogs for the study of nucleic Acid interactions
Abstract
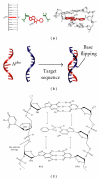
Unnatural nucleosides have been explored to expand the properties and the applications of oligonucleotides. This paper briefly summarizes nucleic acid analogs in which the base is modified or replaced by an unnatural stacking group for the study of nucleic acid interactions. We also describe the nucleoside analogs of a base pair-mimic structure that we have examined. Although the base pair-mimic nucleosides possess a simplified stacking moiety of a phenyl or naphthyl group, they can be used as a structural analog of Watson-Crick base pairs. Remarkably, they can adopt two different conformations responding to their interaction energies, and one of them is the stacking conformation of the nonpolar aromatic group causing the site-selective flipping of the opposite base in a DNA double helix. The base pair-mimic nucleosides can be used to study the mechanism responsible for the base stacking and the flipping of bases out of a nucleic acid duplex.
Figures




Similar articles
-
DNA base flipping by a base pair-mimic nucleoside.Nucleic Acids Res. 2005 Dec 15;33(22):7111-9. doi: 10.1093/nar/gki1018. Print 2005. Nucleic Acids Res. 2005. PMID: 16361269 Free PMC article.
-
Synthesis and properties of the simplified nucleic acid glycol nucleic acid.Acc Chem Res. 2010 Aug 17;43(8):1092-102. doi: 10.1021/ar900292q. Acc Chem Res. 2010. PMID: 20405911
-
Dynamics and energetics of the base flipping conformation studied with base pair-mimic nucleosides.Biochemistry. 2009 Dec 1;48(47):11304-11. doi: 10.1021/bi901496q. Biochemistry. 2009. PMID: 19839646
-
Watson-Crick versus Hoogsteen Base Pairs: Chemical Strategy to Encode and Express Genetic Information in Life.Acc Chem Res. 2021 May 4;54(9):2110-2120. doi: 10.1021/acs.accounts.0c00734. Epub 2021 Feb 16. Acc Chem Res. 2021. PMID: 33591181 Review.
-
Unnatural nucleosides with unusual base pairing properties.Curr Protoc Nucleic Acid Chem. 2001 Aug;Chapter 1:Unit 1.4. doi: 10.1002/0471142700.nc0104s05. Curr Protoc Nucleic Acid Chem. 2001. PMID: 18428819 Review.
Cited by
-
Conformational changes of the phenyl and naphthyl isocyanate-DNA adducts during DNA replication and by minor groove binding molecules.Nucleic Acids Res. 2013 Oct;41(18):8581-90. doi: 10.1093/nar/gkt608. Epub 2013 Jul 19. Nucleic Acids Res. 2013. PMID: 23873956 Free PMC article.
-
Using NMR and molecular dynamics to link structure and dynamics effects of the universal base 8-aza, 7-deaza, N8 linked adenosine analog.Nucleic Acids Res. 2016 Oct 14;44(18):8576-8587. doi: 10.1093/nar/gkw736. Epub 2016 Aug 26. Nucleic Acids Res. 2016. PMID: 27566150 Free PMC article.
-
Chemical Modifications in RNA: Elucidating the Chemistry of dsRNA-Specific Adenosine Deaminases (ADARs).Acc Chem Res. 2023 Sep 19;56(18):2489-2499. doi: 10.1021/acs.accounts.3c00390. Epub 2023 Sep 4. Acc Chem Res. 2023. PMID: 37665999 Free PMC article.
References
-
- Nakano S, Sugimoto N. Energy of nucleic acid self-assemblies: from sequence to function through structure. In: Ariga K, Nalwa HS, et al., editors. Bottom-Up Nanofabrication: Supramolecules, Self-Assemblies, and Organized Films. chapter 8. American Scientific Publishers; 2009. pp. 191–215.
-
- Turner DH, Sugimoto N, Freier SM. RNA structure prediction. Annual Review of Biophysics and Biophysical Chemistry. 1988;17:167–192. - PubMed
-
- Protozanova E, Yakovchuk P, Frank-Kamenetskii MD. Stacked-unstacked equilibrium at the nick site of DNA. Journal of Molecular Biology. 2004;342(3):775–785. - PubMed
-
- Isaksson J, Chattopadhyaya J. A uniform mechanism correlating dangling-end stabilization and stacking geometry. Biochemistry. 2005;44(14):5390–5401. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

