Uridine insertion/deletion editing in trypanosomes: a playground for RNA-guided information transfer
- PMID: 21823228
- PMCID: PMC3154072
- DOI: 10.1002/wrna.82
Uridine insertion/deletion editing in trypanosomes: a playground for RNA-guided information transfer
Abstract
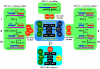
RNA editing is a collective term referring to enzymatic processes that change RNA sequence apart from splicing, 5' capping or 3' extension. In this article, we focus on uridine insertion/deletion mRNA editing found exclusively in mitochondria of kinetoplastid protists. This type of editing corrects frameshifts, introduces start and stops codons, and often adds much of the coding sequence to create an open reading frame. The mitochondrial genome of trypanosomatids, the most extensively studied clade within the order Kinetoplastida, is composed of ∼50 maxicircles with limited coding capacity and thousands of minicircles. To produce functional mRNAs, a multitude of nuclear-encoded factors mediate interactions of maxicircle-encoded pre-mRNAs with a vast repertoire of minicircle-encoded guide RNAs. Editing reactions of mRNA cleavage, U-insertions or U-deletions, and ligation are catalyzed by the RNA editing core complex (RECC, the 20S editosome) while each step of this enzymatic cascade is directed by guide RNAs. These 50-60 nucleotide (nt) molecules are 3' uridylated by RET1 TUTase and stabilized via association with the gRNA binding complex (GRBC). Remarkably, the information transfer between maxicircle and minicircle transcriptomes does not rely on template-dependent polymerization of nucleic acids. Instead, intrinsic substrate specificities of key enzymes are largely responsible for the fidelity of editing. Conversely, the efficiency of editing is enhanced by assembling enzymes and RNA binding proteins into stable multiprotein complexes. WIREs RNA 2011 2 669-685 DOI: 10.1002/wrna.82 For further resources related to this article, please visit the WIREs website.
Copyright © 2011 John Wiley & Sons, Ltd.
Figures

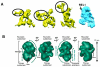


References
-
- Benne R, Van den Burg J, Brakenhoff J, Sloof P, Van Boom J, Tromp M. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. - PubMed
-
- Feagin JE, Abraham J, Stuart K. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell. 1988;53:413–422. - PubMed
-
- Shaw J, Feagin JE, Stuart K, Simpson L. Editing of mitochondrial mRNAs by uridine addition and deletion generates conserved amino acid sequences and AUG initiation codons. Cell. 1988;53:401–411. - PubMed
-
- Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. - PubMed
-
- Sturm NR, Simpson L. Kinetoplast DNA minicircles encode guide RNAs for editing of cytochrome oxidase subunit III mRNA. Cell. 1990;61:879–884. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

