Discovery of a cytokinin deaminase
- PMID: 21823622
- PMCID: PMC3199332
- DOI: 10.1021/cb200198c
Discovery of a cytokinin deaminase
Abstract
An enzyme of unknown function within the amidohydrolase superfamily was discovered to catalyze the hydrolysis of N-6-substituted adenine derivatives, several of which are cytokinins. Cytokinins are a common type of plant hormone and N-6-substituted adenines are also found as modifications to tRNA. Patl2390, from Pseudoalteromonas atlantica T6c, was shown to hydrolytically deaminate N-6-isopentenyladenine to hypoxanthine and isopentenylamine with a k(cat)/K(m) of 1.2 × 10(7) M(-1) s(-1). Additional substrates include N-6-benzyl adenine, cis- and trans-zeatin, kinetin, O-6-methylguanine, N-6-butyladenine, N-6-methyladenine, N,N-dimethyladenine, 6-methoxypurine, 6-chloropurine, and 6-thiomethylpurine. This enzyme does not catalyze the deamination of adenine or adenosine. A comparative model of Patl2390 was computed using the three-dimensional crystal structure of Pa0148 (PDB code 3PAO ) as a structural template, and docking was used to refine the model to accommodate experimentally identified substrates. This is the first identification of an enzyme that will hydrolyze an N-6-substituted side chain larger than methylamine from adenine.
Figures


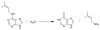
Similar articles
-
Pa0148 from Pseudomonas aeruginosa catalyzes the deamination of adenine.Biochemistry. 2011 Aug 2;50(30):6589-97. doi: 10.1021/bi200868u. Epub 2011 Jul 6. Biochemistry. 2011. PMID: 21710971 Free PMC article.
-
Enzymatic deamination of the epigenetic base N-6-methyladenine.J Am Chem Soc. 2011 Feb 23;133(7):2080-3. doi: 10.1021/ja110157u. Epub 2011 Jan 28. J Am Chem Soc. 2011. PMID: 21275375 Free PMC article.
-
Topolins and hydroxylated thidiazuron derivatives are substrates of cytokinin O-glucosyltransferase with position specificity related to receptor recognition.Plant Physiol. 2005 Mar;137(3):1057-66. doi: 10.1104/pp.104.057174. Epub 2005 Feb 22. Plant Physiol. 2005. PMID: 15728338 Free PMC article.
-
The Hulks and the Deadpools of the Cytokinin Universe: A Dual Strategy for Cytokinin Production, Translocation, and Signal Transduction.Biomolecules. 2021 Feb 3;11(2):209. doi: 10.3390/biom11020209. Biomolecules. 2021. PMID: 33546210 Free PMC article. Review.
-
Naturally Occurring and Artificial N9-Cytokinin Conjugates: From Synthesis to Biological Activity and Back.Biomolecules. 2020 May 29;10(6):832. doi: 10.3390/biom10060832. Biomolecules. 2020. PMID: 32485963 Free PMC article. Review.
Cited by
-
Cytidine deaminases catalyze the conversion of N(S,O)4-substituted pyrimidine nucleosides.Sci Adv. 2023 Feb 3;9(5):eade4361. doi: 10.1126/sciadv.ade4361. Epub 2023 Feb 3. Sci Adv. 2023. PMID: 36735785 Free PMC article.
-
Prediction of substrates for glutathione transferases by covalent docking.J Chem Inf Model. 2014 Jun 23;54(6):1687-99. doi: 10.1021/ci5001554. Epub 2014 May 16. J Chem Inf Model. 2014. PMID: 24802635 Free PMC article.
-
Function discovery and structural characterization of a methylphosphonate esterase.Biochemistry. 2015 May 12;54(18):2919-30. doi: 10.1021/acs.biochem.5b00199. Epub 2015 Apr 28. Biochemistry. 2015. PMID: 25873441 Free PMC article.
-
Manipulation of cytokinin level in the ergot fungus Claviceps purpurea emphasizes its contribution to virulence.Curr Genet. 2018 Dec;64(6):1303-1319. doi: 10.1007/s00294-018-0847-3. Epub 2018 May 30. Curr Genet. 2018. PMID: 29850931
-
Deamination of 6-aminodeoxyfutalosine in menaquinone biosynthesis by distantly related enzymes.Biochemistry. 2013 Sep 17;52(37):6525-36. doi: 10.1021/bi400750a. Epub 2013 Sep 4. Biochemistry. 2013. PMID: 23972005 Free PMC article.
References
-
- Seibert CM, Raushel FM. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry. 2005;44:6383–6391. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

