Clustering of Alpers disease mutations and catalytic defects in biochemical variants reveal new features of molecular mechanism of the human mitochondrial replicase, Pol γ
- PMID: 21824913
- PMCID: PMC3241644
- DOI: 10.1093/nar/gkr618
Clustering of Alpers disease mutations and catalytic defects in biochemical variants reveal new features of molecular mechanism of the human mitochondrial replicase, Pol γ
Abstract
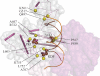
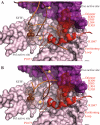
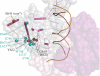
Mutations in Pol γ represent a major cause of human mitochondrial diseases, especially those affecting the nervous system in adults and in children. Recessive mutations in Pol γ represent nearly half of those reported to date, and they are nearly uniformly distributed along the length of the POLG1 gene (Human DNA Polymerase gamma Mutation Database); the majority of them are linked to the most severe form of POLG syndrome, Alpers-Huttenlocher syndrome. In this report, we assess the structure-function relationships for recessive disease mutations by reviewing existing biochemical data on site-directed mutagenesis of the human, Drosophila and yeast Pol γs, and their homologs from the family A DNA polymerase group. We do so in the context of a molecular model of Pol γ in complex with primer-template DNA, which we have developed based upon the recently solved crystal structure of the apoenzyme form. We present evidence that recessive mutations cluster within five distinct functional modules in the catalytic core of Pol γ. Our results suggest that cluster prediction can be used as a diagnosis-supporting tool to evaluate the pathogenic role of new Pol γ variants.
Figures




Similar articles
-
Disease mutations in the human mitochondrial DNA polymerase thumb subdomain impart severe defects in mitochondrial DNA replication.J Biol Chem. 2009 Jul 17;284(29):19501-10. doi: 10.1074/jbc.M109.011940. Epub 2009 May 28. J Biol Chem. 2009. PMID: 19478085 Free PMC article.
-
Depletion of mitochondrial DNA in fibroblast cultures from patients with POLG1 mutations is a consequence of catalytic mutations.Hum Mol Genet. 2008 Aug 15;17(16):2496-506. doi: 10.1093/hmg/ddn150. Epub 2008 May 16. Hum Mol Genet. 2008. PMID: 18487244 Free PMC article.
-
Yeast cells expressing the human mitochondrial DNA polymerase reveal correlations between polymerase fidelity and human disease progression.J Biol Chem. 2014 Feb 28;289(9):5970-85. doi: 10.1074/jbc.M113.526418. Epub 2014 Jan 7. J Biol Chem. 2014. PMID: 24398692 Free PMC article.
-
Consequences of mutations in human DNA polymerase gamma.Gene. 2005 Jul 18;354:125-31. doi: 10.1016/j.gene.2005.03.029. Gene. 2005. PMID: 15913923 Review.
-
DNA polymerase gamma and mitochondrial disease: understanding the consequence of POLG mutations.Biochim Biophys Acta. 2009 May;1787(5):312-9. doi: 10.1016/j.bbabio.2008.10.007. Epub 2008 Oct 29. Biochim Biophys Acta. 2009. PMID: 19010300 Free PMC article. Review.
Cited by
-
What is influencing the phenotype of the common homozygous polymerase-γ mutation p.Ala467Thr?Brain. 2012 Dec;135(Pt 12):3614-26. doi: 10.1093/brain/aws298. Brain. 2012. PMID: 23250882 Free PMC article.
-
Alpers disease mutations in human DNA polymerase gamma cause catalytic defects in mitochondrial DNA replication by distinct mechanisms.Front Genet. 2015 Apr 9;6:135. doi: 10.3389/fgene.2015.00135. eCollection 2015. Front Genet. 2015. PMID: 25914719 Free PMC article.
-
Animal Mitochondrial DNA Replication.Enzymes. 2016;39:255-92. doi: 10.1016/bs.enz.2016.03.006. Epub 2016 May 9. Enzymes. 2016. PMID: 27241933 Free PMC article. Review.
-
Mitochondrial DNA maintenance in Drosophila melanogaster.Biosci Rep. 2022 Nov 30;42(11):BSR20211693. doi: 10.1042/BSR20211693. Biosci Rep. 2022. PMID: 36254835 Free PMC article. Review.
-
Reduced stimulation of recombinant DNA polymerase γ and mitochondrial DNA (mtDNA) helicase by variants of mitochondrial single-stranded DNA-binding protein (mtSSB) correlates with defects in mtDNA replication in animal cells.J Biol Chem. 2011 Nov 25;286(47):40649-58. doi: 10.1074/jbc.M111.289983. Epub 2011 Sep 26. J Biol Chem. 2011. PMID: 21953457 Free PMC article.
References
-
- Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Ann. Rev. Biochem. 2004;73:293–320. - PubMed
-
- Graziewicz MA, Longley MJ, Copeland WC. DNA polymerase gamma in mitochondrial DNA replication and repair. Chem. Rev. 2006;106:383–405. - PubMed
-
- Yakubovskaya E, Chen Z, Carrodeguas JA, Kisker C, Bogenhagen DF. Functional human mitochondrial DNA polymerase gamma forms a heterotrimer. J. Biol. Chem. 2006;281:374–382. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

