Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly
- PMID: 21825076
- PMCID: PMC3153650
- DOI: 10.1083/jcb.201012063
Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly
Abstract
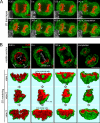
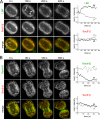
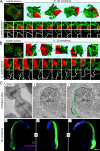
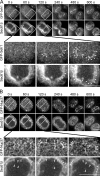
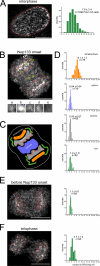
During mitosis, the nuclear envelope merges with the endoplasmic reticulum (ER), and nuclear pore complexes are disassembled. In a current model for reassembly after mitosis, the nuclear envelope forms by a reshaping of ER tubules. For the assembly of pores, two major models have been proposed. In the insertion model, nuclear pore complexes are embedded in the nuclear envelope after their formation. In the prepore model, nucleoporins assemble on the chromatin as an intermediate nuclear pore complex before nuclear envelope formation. Using live-cell imaging and electron microscope tomography, we find that the mitotic assembly of the nuclear envelope primarily originates from ER cisternae. Moreover, the nuclear pore complexes assemble only on the already formed nuclear envelope. Indeed, all the chromatin-associated Nup107-160 complexes are in single units instead of assembled prepores. We therefore propose that the postmitotic nuclear envelope assembles directly from ER cisternae followed by membrane-dependent insertion of nuclear pore complexes.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

